Theme
Chromosomes are the blueprint of life and a platform of many essential functions, including DNA
replication, transcription, recombination, repair, and segregation. On chromosomes, multiple DNA-protein
interactions are coordinated into networks, which play essential roles in controlling chromosome
shape and ensuring faithful execution of various activities on DNA. Our main challenge is to understand
the function and structure of chromosomes at every level.
About Research
Challenge to solve the enigma of structure and function of chromosome
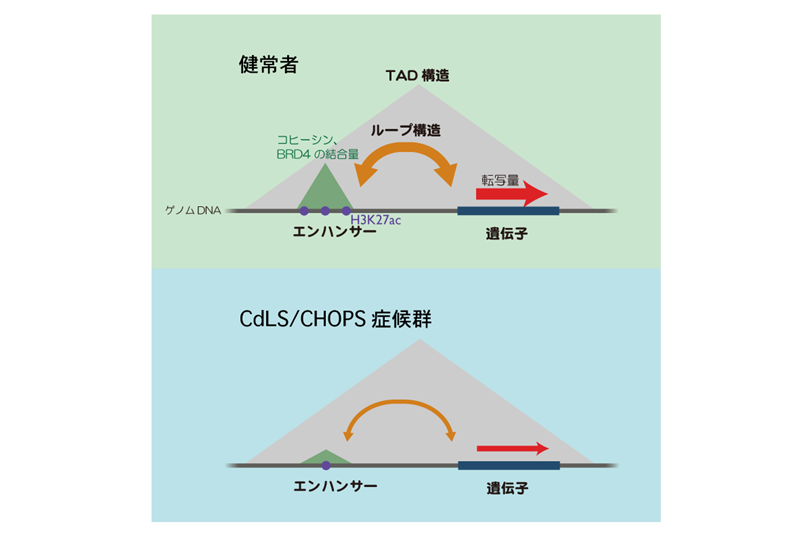
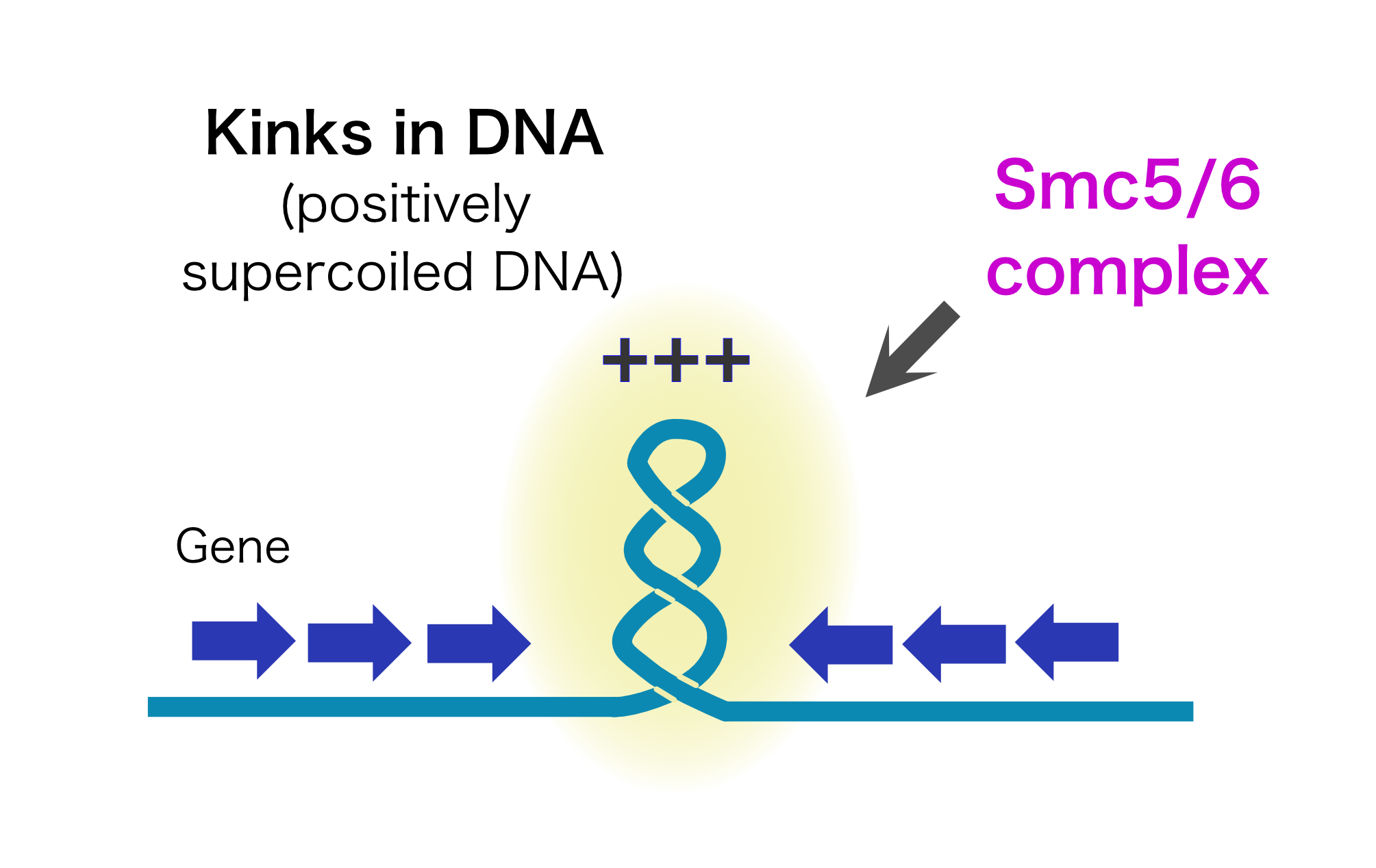
Chromosome is a blueprint of life. For the survival and proliferation of all living organisms, it is essential that chromosomes (= genetic information) are succeeded to next generation. Chromosome is a genetic platform where hundreds of proteins play their own roles for the maintenance and expression of genetic information. Our laboratory is one of the leading laboratories which has succeeded in clarifying the elementary process and interplay of various chromosomal functions using genomic methods. Currently, we are trying to analyze the regulatory mechanism of the function and structure of a huge human chromosomes. In particular, our focus is to understand the role of protein complex called cohesin in organizing the chromosome 3D structure and function. Through the analysis of cohesin complex, we are trying to elucidate the actual state of huge transcription structure (enhanceosome) whose entire picture is unknown. For this, in addition to cutting-edge genomic technologies, we are also developing a transcriptional reconstitution system in vitro using synthetic DNA of hundreds of kilo base pairs in size. We also analyze chromosome structure and functions of cells derived from a series of human rare disease patients caused by mutations in genes responsible for organizing high order chromosome structure. By combining these unique strategies, we are eager to persuit regulatory mechanisms that link chromosome structure and function at every level.
Publication
- Minamino M, Tei S, Negishi L, Kanemaki M, Yoshimura A, Sutani T, *Bando M, * Shirahige K (2018) Temporal Regulation of ESCO2 Degradation by the MCM Complex, the CUL4-DDB1-VPRBP Complex, and the Anaphase Promoting Complex Curr Biol. 28:2665-2672(*Shared Corresponding Authors)
- Sutani T, Sakata T, Nakato R, Masuda K, Ishibashi1 M, Yamashita D, Suzuki Y, Hirano T, Bando M, Shirahige K. (2015) Condensin targets and reduces unwound DNA structure associated with transcription in mitotic chromosome condensation. Nat Commun 6:7815.
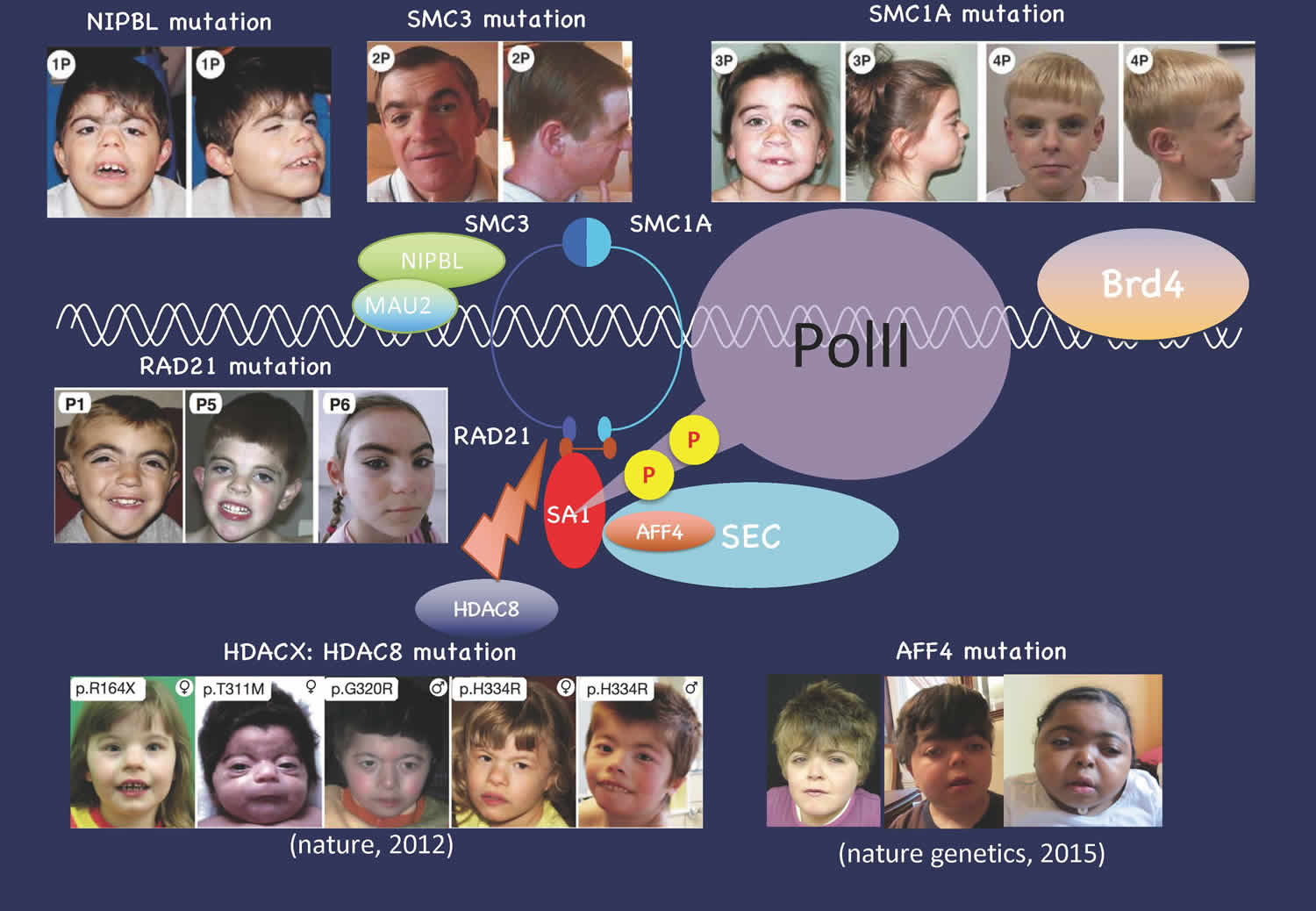
- Izumi K, Nakato R, Zhang Z, Edmondson AC, Noon S, Dulik MC, Rajagopalan R, Venditti CP, Gripp K, Samanich J, Zackai EH, Deardorff MA, Clark D, Allen JL, Dorsett D, Misulovin Z, Komata M, Bando M, Kaur M, Katou Y, *Shirahige K, *Krantz ID. (2015) Germline gain-of-function mutations in AFF4 cause a developmental syndrome functionally linking the super elongation complex and cohesin. Nat Genet. 47:338-44. (*shared corresponding authors)
- Deardorff MA, Bando M, Nakato R, Watrin E, Itoh T, Minamino M, Saitoh K, Komata M, Katou Y, Clark D, Cole KE, De Baere E, Decroos C, Di Donato N, Ernst S, Francey LJ, Gyftodimou Y, Hirashima K, Hullings M, Ishikawa Y, Jaulin C, Kaur M, Kiyono T, Lombardi PM, Magnaghi-Jaulin L, Mortier GR, Nozaki N, Petersen MB, Seimiya H, Siu VM, Suzuki Y, Takagaki K, Wilde JJ, Willems PJ, Prigent C, Gillessen-Kaesbach G, Christianson DW, Kaiser FJ, Jackson LG, Hirota T, Krantz ID, Shirahige K. (2012) HDAC8 mutations in Cornelia de Lange syndrome affect the cohesin acetylation cycle. Nature. 489:313-7.
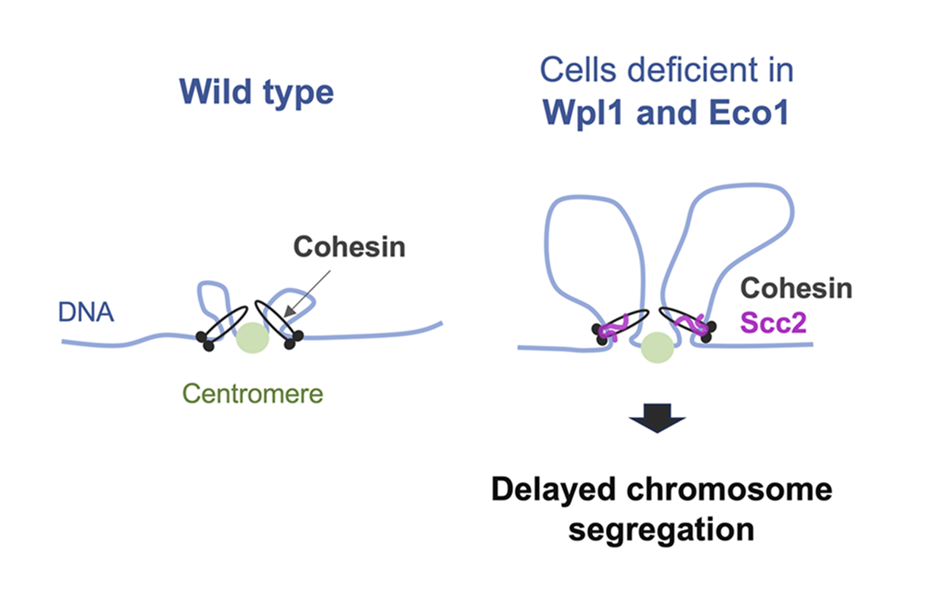
- Sutani T, Kawaguchi T, Kanno R, Itoh T, Shirahige K. (2009) Budding yeast Wpl1(Rad61)-Pds5 complex counteracts sister chromatid cohesion-establishing reaction. Curr Biol. 19:492-497.
- K. S. Wendt*, K. Yoshida*, T. Itoh*, M. Bando, B. Koch, E. Schirghuber, S. Tsutsumi, G. Nagae, K. Ishihara, T. Mishiro, K. Yahata, F. Imamoto, H. Aburatani, M. Nakao, N. Imamoto, K. Maeshima, K. Shirahige#, and J.-M. Peters# (2008) Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature (article). 451, 796-801 (*equally contributed author) (#shared corresponding authors)
- A. Lengronne, Y. Katou, S. Mori, S. Yokobayashi, G. Kelly, T. Itoh, Y. Watanabe, K. Shirahige*, and F. Uhlmann* (2004) Cohesin relocation from sites of chromosomal loading to places of convergent transcriptions. Nature, 430, 573-578 (*equally contributed authors)
- Y. Katou, Y. Kanoh, M. Bando, H. Noguchi, H. Tanaka, T. Ashikari, K. Sugimoto, and K. Shirahige (2003) S-phase checkpoint proteins Tof1 and Mrc1 form a stable replication pausing complex. Nature, 424, 1078-1083
- K. Shirahige, Y. Hori, K. Shiraishi, M. Yamashita, K. Takahashi, C. Obuse, T. Tsurimoto and H. Yoshikawa (1998) Regulation of DNA-replication origins during cell-cycle progression. Nature, 395, 618-621
- K. Shirahige, T. Iwasaki, MB. Rashid, N. Ogasawara and H. Yoshikawa (1993) Location and characterization of autonomously replicating sequences from chromosome VI of Saccharomyces cerevisiae. Mol Cell Biol., 13, 5043-5056
Katsuhiko Shirahige
Professor
Ph.D.
Graduate School of Medicine, Graduate School of Arts and Sciences
Takashi Sutani
Associate Professor
Ph.D.
Graduate School of Medicine
Yuri Nakagawa
Project Associate Professor
Ph.D.
Masashige Bando
Lecturer
Ph.D.
Graduate School of Medicine
Katsunori Fujiki
Research Associate
Ph.D.
Toyonori Sakata
Research Associate
Ph. D.