Theme
Genome is the most important information on the earth that determines the design of life while the material DNA is quite “fragile”. We try to reveal the mechanism of genome maintenance and the
relationship to cellular senescence and tumorigenesis.
About Research
Genome maintenance, Cellular senescence and Tumorigenesis.
How is the genome integrity maintained?
Human genome has ~ 3 billions base pairs, corresponding to ~ 2 m in length. Such a long DNA is packed into the small nucleus (~ 5 μm diameters) and duplicated in mitosis. It is easy to imagine that the genome is entangled and broken in the small compartment. In addition, the genome gets damaged by ROS, UV and chemical modifi cations. Seriously damaged genome induces apoptosis to kill the cell. Slight damaged one activates the DNA repair system. But the some lesions on the genome may escape from the system and accumulate. The accumulation reduces cellular functions and induces senescence. It may cause cancer, too. As a human body has ~ 4 trillions cells, it is a quite tough job to maintain the genome integrity in all of them. Especially, long-life cells, such as germ line and stem cells need intensive care. We study how those cells regenerate genome and keep the integrity.
What is the“ Aging signal” from damaged genome?
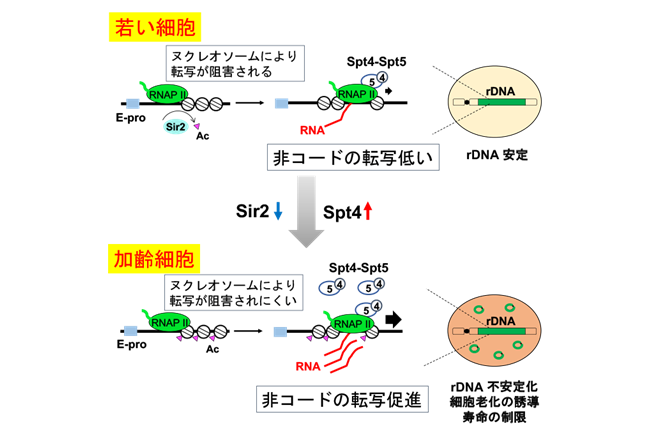
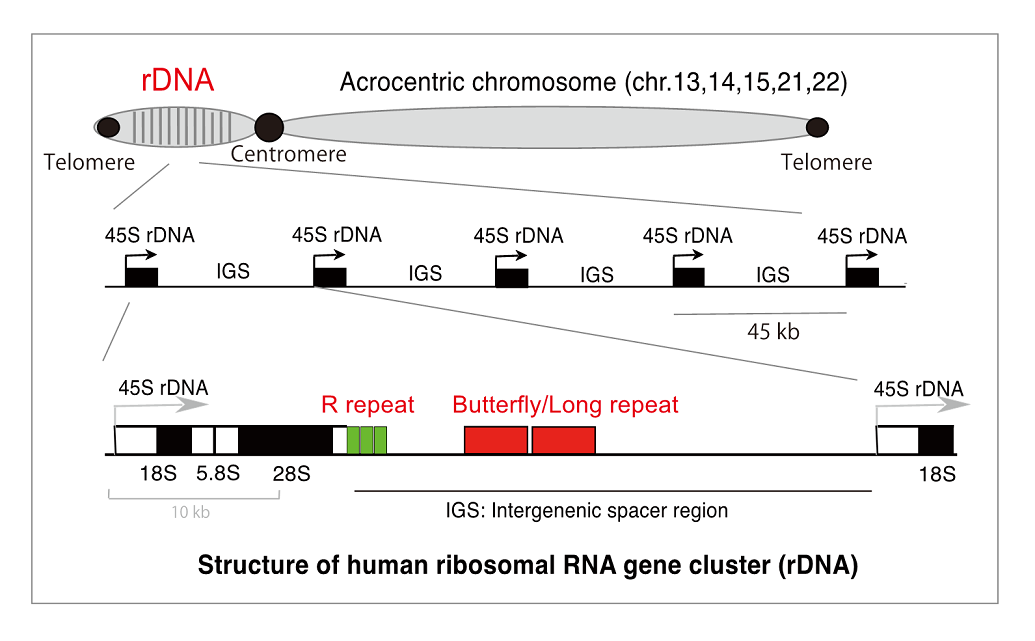
In eukaryotic cells, the genome has several “fragile” sites that are especially unstable. The ribosomal RNA gene repeat (rDNA) is the biggest one. The repeat occupies ~ 10% of yeast and ~ 0.5 % of human genome. Recently, we found that rDNA stability determines yeast lifespan. Therefore, we speculate that the rDNA is a major source of “Aging signal” that induces cellular senescence and restricts the lifespan. We would like to identify the signal.
Publication
- Ribosomal RNA gene repeats, their stability and cellular senescence.
Kobayashi T.
Proc Jpn Acad Ser B Phys Biol Sci. 2014 Apr 11;90(4): 119-129. doi: 10.2183/pjab.90.119.
- Cellular Senescence in Yeast Is Regulated by rDNA Noncoding Transcription.
Saka K, Ide S, Ganley AR, Kobayashi T.
Curr Biol. 2013 Sep 23;23(18):1794-8. doi: 10.1016/j.cub.2013.07.048.
- Regulation of ribosomal RNA gene copy number and its role in modulating genome integrity and evolutionary adaptability in yeast.
Kobayashi T.
Cell Mol Life Sci. 2011 Apr;68(8):1395-403. doi: 10.1007/s00018-010-0613-2.
- Abundance of ribosomal RNA gene copies maintains genome integrity.
Ide S, Miyazaki T, Maki H, Kobayashi T.
Science. 2010 Feb 5;327(5966):693-6. doi: 10.1126/science.1179044.
- The effect of replication initiation on gene amplification in the rDNA and its relationship to aging.
Ganley ARD, Ide S, Saka K, Kobayashi T.
Mol Cell. 2009 Sep 11;35(5):683-93. doi: 10.1016/j.molcel.2009.07.012.
- Recombination regulation by transcription-induced cohesin dissociation in rDNA repeats.
Kobayashi T and Ganley ARD.
Science. 2005 Sep 2;309(5740):1581-4. doi: 10.1126/science.1116102.
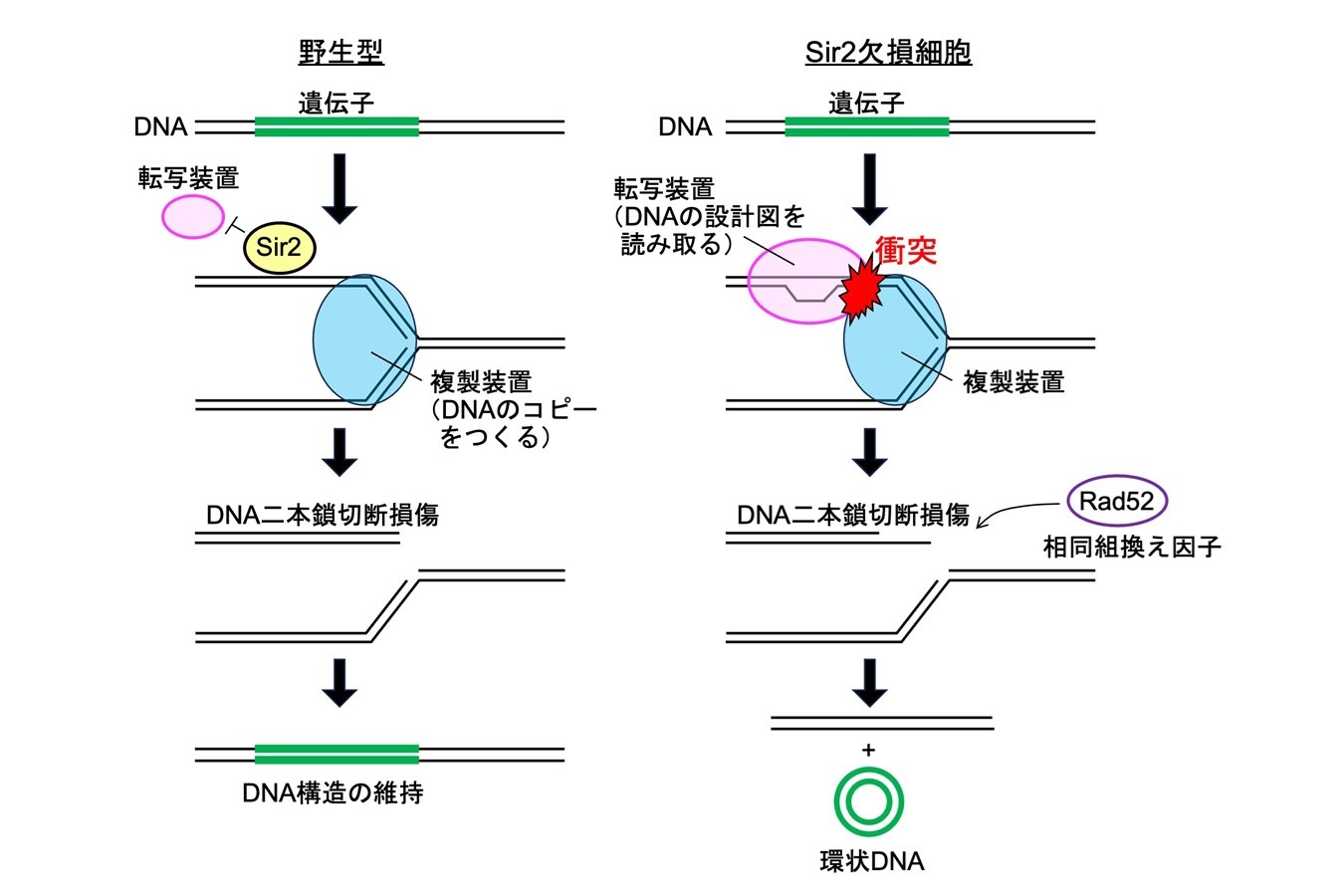
- SIR2 regulates recombination between different rDNA repeats, but not recombination within individual rRNA genes in yeast.
Kobayashi T, Horiuchi T, Tongaonkar P, Vu L, Nomura M .
Cell. 2004 May 14;117(4):441-53. doi: 10.1016/S0092-8674(04)00414-3
- Transcription-dependent recombination and the role of fork collision in yeast rDNA.
Takeuchi Y, Horiuchi T, Kobayashi T.
Genes Dev. 2003 Jun 15;17(12):1497-506. doi: 10.1101/gad.1085403
- Expansion and contraction of ribosomal DNA repeats in Saccharomyces cerevisiae: requirement of replication fork blocking (Fob1) protein and the role of RNA polymerase I.
Kobayashi T, Heck DJ, Nomura M, Horiuchi T.
Genes Dev. 1998 Dec 15;12(24):3821-30. doi:10.1101/gad.12.24.3821
- Evidence of a ter specific binding protein essential for the termination reaction of DNA replication in Escherichia coli.
Kobayashi T, Hidaka M, Horiuchi T.
EMBO J. 1989 Aug;8(8):2435-41..
Takehiko Kobayashi
Professor
Ph.D.
Graduate School of Science
Eriko Oya
Research Associate
Ph.D.
Yutaro Hori
Research Associate
Ph.D.
Shuichi Yanagi
Project Research Associate
Ph.D.
Tomoko Asakura
Technical Specialist