Theme
Non-coding RNAs do not serve as the template for protein synthesis but they themselves act as functional molecules that regulate complex biological processes. By combining various methodologies, we aim at dissecting the molecular mechanism of non-coding RNAs.
About Research
Understanding diverse RNA functions and mechanisms
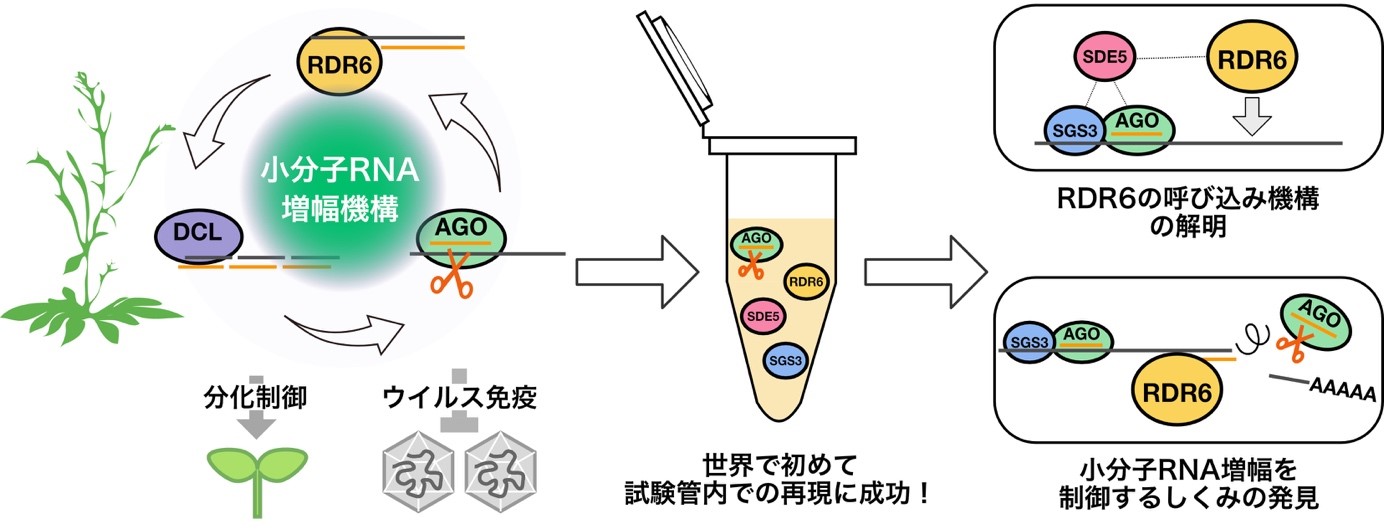
Most genetic information encoded by the genomic DNA is first transcribed as messenger RNAs (mRNAs), followed by translation to proteins to exert their functions. However, recent studies have revealed that cells express not only mRNAs but numerous non-coding RNAs (ncRNAs), which act as functional molecules without being translated to proteins. For instance, small RNAs of 20–30 nucleotides long, including microRNAs, siRNAs and piRNAs, negatively regulate their complementary target RNAs, thereby controlling complex biological processes. Moreover, small RNAs are utilized as a useful tool in basic biology and more recently as medical drugs. Yet, our knowledge on production and function of these ncRNA species is still very limited. We are challenging this new frontier of the RNA world by combining biochemistry, biophysics, and cellular and developmental biology.
Publication
- Iruka Eliminates Dysfunctional Argonaute by Selective Ubiquitination of Its Empty State.
*Kobayashi H, Shoji K, Kiyokawa K, Negishi L, *Tomari Y.
Mol Cell. 2019 Jan 3;73(1):119-129.e5.
- Conformational activation of Argonaute by distinct yet coordinated actions of the Hsp70 and Hsp90 chaperone systems.
Tsuboyama K, *Tadakuma H, *Tomari Y.
Mol Cell. 2018 May 17;70(4):722-729.e4.
- Silencing messages in a unique way.
*Iwakawa HO, *Tomari Y.
Nat Plants. 2017 Oct;3(10):769-770.
- The poly(A) tail blocks RDR6 from converting self mRNAs into substrates for gene silencing.
Baeg K, *Iwakawa HO, *Tomari Y.
Nat Plants. 2017 Mar 20;3:17036.
- Codon Usage and 3' UTR Length Determine Maternal mRNA Stability in Zebrafish.
*Mishima Y, Tomari Y.
Mol Cell. 2016 Mar 17;61(6):874-85.
- Identification and functional analysis of the pre-piRNA 3′ Trimmer in silkworms.
Izumi N, Shoji K, Sakaguchi Y, Honda S, Kirino Y, Suzuki T, Katsuma S, *Tomari Y.
Cell. 2016 Feb 25;164(5):962-73.
- Single-molecule analysis of the target cleavage reaction by Drosophila RNAi enzyme complex.
Yao C, Sasaki HM, Ueda T, *Tomari Y, *Tadakuma H.
Mol Cell. 2015 Jul 2;59(1):125-32.
- Defining fundamental steps in the assembly of the Drosophila RNAi enzyme complex.
Iwasaki S, Sasaki HM, Sakaguchi Y, Suzuki T, *Tadakuma H, *Tomari Y.
Nature. 2015 May 28;521(7553):533-6.
- The initial uridine of primary piRNAs does not create the tenth adenine that is the hallmark of secondary piRNAs.
Wang W, Yoshikawa M, Han BW, Izumi N, *Tomari Y, *Weng Z, *Zamore PD.
Mol Cell. 2014 Dec 4;56(5):708-16.
- microRNAs block assembly of eIF4F translation initiation complex in Drosophila.
Fukaya T, Iwakawa HO, *Tomari Y.
Mol Cell. 2014 Oct 2;56(1):67-78.
Yukihide Tomari
Professor
Ph.D.
Graduate School of Frontier Sciences
Fumiko Kawasaki
Research Associate
Ph.D.
Hirono Kina
Research Associate
Ph.D.
Saori Shinoda
Research Associate
Ph.D.
Natsuko Izumi
Technical Specialist
Ph.D.