Theme
Our laboratory is focused on understanding the mechanisms by which eukaryotic chromatin regulates gene expression
and genomic stability, which are fundamental to life. To do so, we perform biochemical, molecular biological, and
structural biological studies of in vitro reconstituted chromatin.
About Research
The aim of our laboratory is to reveal how chromatin regulates the fundamental processes of life. We do this by using biochemical and structural biological approaches as our primary tools to study in vitro reconstituted chromatin.
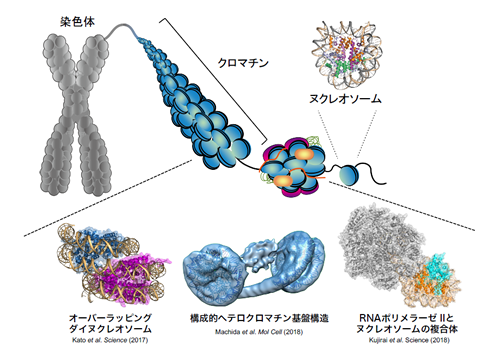
In eukaryotic cells, genomic DNA is accommodated within the nucleus.
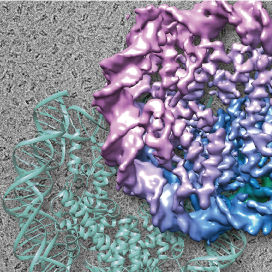
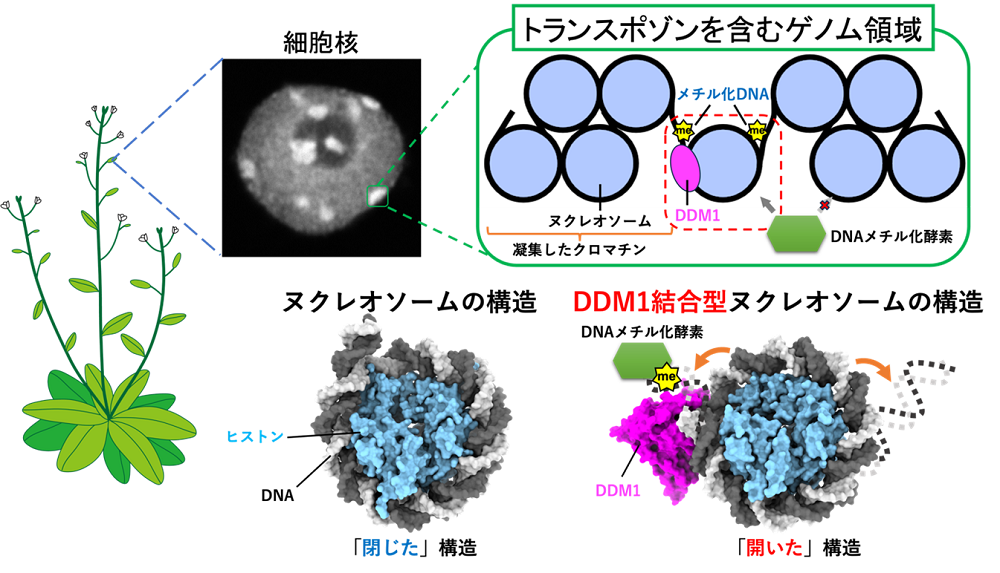
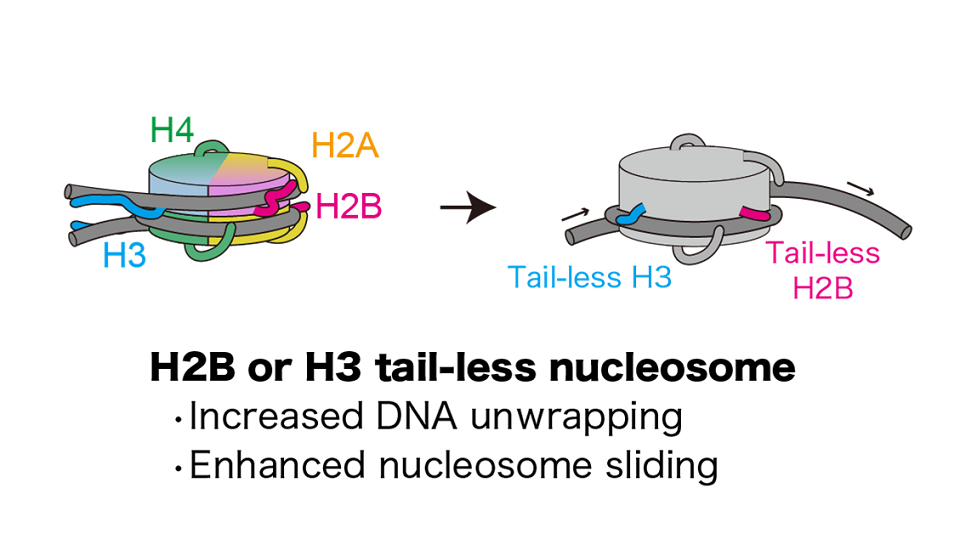
In a human cell, the length of the genomic DNA is about 2 meters. A 150 base-pair segment of genomic DNA is wrapped around an octameric histone protein core composed of two copies of each of four histones H2A, H2B, H3, and H4. This histone-DNA complex is called “nucleosome”, which is the basic unit of chromatin. The nucleosomes are spaced along the genomic DNA, forming a “beads-on-a-string” topology. An array of nucleosomes interacts with various chromatin-associating proteins and RNAs, creating a dynamic higher-order chromatin structure.
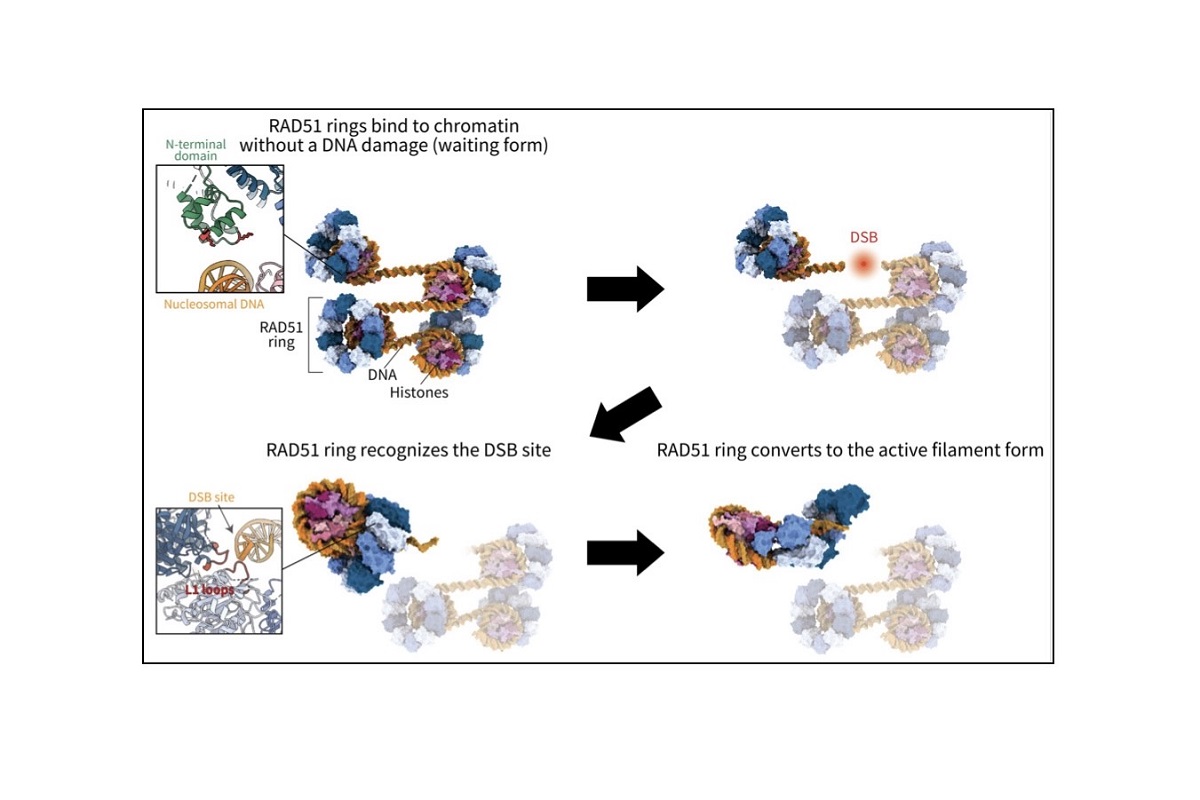
The higher-order structure of chromatin is intimately involved in the regulation of gene expression. Our research is focused on elucidating unresolved mechanisms by which chromatin regulates gene expression. Our main strategy is to reconstitute chromatin in vitro, which serves as a basis for studying various mechanisms that take place on the chromatin. The mechanisms are studied using various biochemical assays and structure determination methods such as X-ray crystallography and cryo-electron microscopy. We developed technologies that allow us to reconstitute high quality chromatin in vitro. We have been successful in analyzing diverse forms of chromatin, such as nucleosomes localized specifically at transcription start sites, nucleosomes found in cancer cells, chromatin composed of multiple nucleosomes, and nucleosomes bound to various nuclear proteins including transcription factors and linker histones, etc. Our laboratory is constantly refining the technologies in order to reconstitute chromatin that closely mimics the in vivo state.
Publication
- Hatazawa S, Liu J, Takizawa Y, Zandian M, Negishi L, Kutateladze TG, Kurumizaka H, Structural basis for binding diversity of acetyltransferase p300 to the nucleosome (2022) iScience, doi: 10.1016/j.isci.2022.104563.
- Hirai S, Tomimatsu K, Miyawaki-Kuwakado A, Takizawa Y, Komatsu T, Tachibana T, Fukushima Y, Takeda Y, Negishi L, Kujirai T, Koyama M, Ohkawa Y, Kurumizaka H, Unusual nucleosome formation and transcriptome influence by the histone H3mm18 variant (2022) Nucleic Acids Res, doi: 10.1093/nar/gkab1137.
- Hirano R, Arimura Y, Kujirai T, Shibata M, Okuda A, Morishima K, Inoue R, Sugiyama M, Kurumizaka H, Histone variant H2A.B-H2B dimers are spontaneously exchanged with canonical H2A-H2B in the nucleosome (2021) Commun Biol, doi: 10.1038/s42003-021-01707-z.
- Sato S, Takizawa Y, Hoshikawa F, Dacher M, Tanaka H, Tachiwana H, Kujirai T, Iikura Y, Ho CH, Adachi N, Patwal I, Flaus A, Kurumizaka H, Cryo-EM structure of the nucleosome core particle containing Giardia lamblia histones (2021) Nucleic Acids Res, doi: 10.1093/nar/gkab644.
- Tanaka H, Takizawa Y, Takaku M, Kato D, Kumagawa Y, Grimm SA, Wade PA, Kurumizaka H. Interaction of the pioneer transcription factor GATA3 with nucleosomes (2020) Nat Commun, doi: 10.1038/s41467-020-17959-y.
- Kujirai T, Zierhut C, Takizawa Y, Kim R, Negishi L, Uruma N, Hirai S, Funabiki H, Kurumizaka H, Structural basis for the inhibition of cGAS by nucleosomes (2020) Science, doi: 10.1126/science.abd0237.
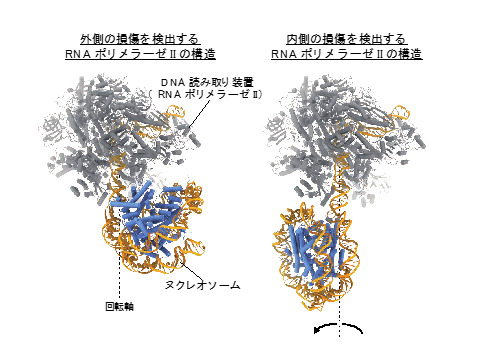
- Ehara H, Kujirai T, Fujino Y, Shirouzu M, Kurumizaka H, Sekine SI, Structural insight into nucleosome transcription by RNA polymerase II with elongation factors. (2019) Science, doi: 10.1126/science.aav8912.
- Kujirai T, Ehara H, Fujino Y, Shirouzu M, Sekine SI, Kurumizaka H, Structural basis of the nucleosome transition during RNA polymerase II passage (2018) Science, doi: 10.1126/science.aau9904.
- Machida S, Takizawa Y, Ishimaru M, Sugita Y, Sekine S, Nakayama JI, Wolf M, Kurumizaka H, Structural Basis of Heterochromatin Formation by Human HP1 (2018) Mol. Cell 69(3):385-397.e8. doi: 10.1016/j.molcel.2017.12.011.
- Kato D, Osakabe A, Arimura Y, Mizukami Y, Horikoshi N, Saikusa K, Akashi S, Nishimura Y, Park SY, Nogami J, Maehara K, Ohkawa Y, Matsumoto A, Kono H, Inoue R, Sugiyama M, Kurumizaka H, Crystal structure of the overlapping dinucleosome composed of hexasome and octasome (2017) Science, 356(6334):205-208. doi: 10.1126/science.aak9867
Hitoshi Kurumizaka
Professor
Ph.D.
Graduate School of Science
Yukimasa Takizawa
Associate Professor
Ph.D.
Naoki Horikoshi
Associate Professor
Ph.D.
Tomoya Kujirai
Research Associate
Ph.D.
Syota Matsumoto
Research Associate
Ph.D.
Shoko Sato
Research Associate
Ph. D.