Theme
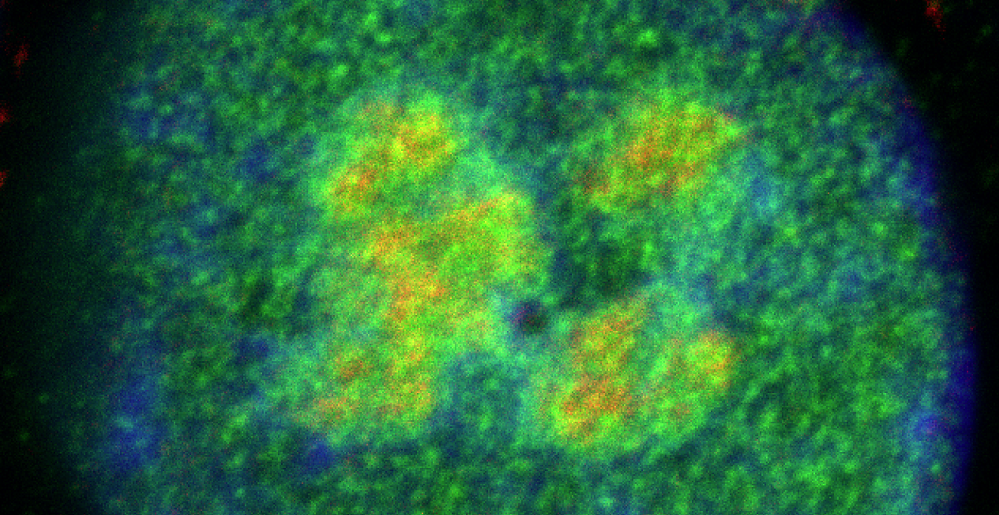
Recent discoveries using cutting-edge imaging techniques have begun to reveal the dynamics of biomolecules inside the cell. Our laboratory investigates how these dynamics can be modulated to control enzymatic activity and potentiate oncogenic signalling, with the ultimate goal of developing therapeutic agents to counteract the drivers of cancer.
About Research
Understanding the molecular grammar underlying protein diffusivity in cancer cells
DNA binding-independent functions of transcription factors
While oncogenic transcription factors are known to stimulate gene expression through DNA binding, evidence suggests they may also enhance transcription through DNA binding-independent mechanisms. This feature of transcription factors is under-researched, but new insights are beginning to demonstrate how the molecular grammar of these proteins (i.e. the biophysical properties imparted by their amino acid composition) can influence protein localization, condensation, diffusivity and enzymatic activity. We want to explore these alternate pathways using in vitro biochemical techniques and next-generation sequencing approaches to reveal new modalities for therapeutic targeting of oncoproteins.
Modulating protein kinetics in the cancer cell
State-of-the-art techniques have revealed that biomolecule kinetics varies significantly across the cell. Many different biophysical features likely control cellular diffusivity. For example, transcription condensate form on DNA creating highly concentrated protein compartments where protein kinetics are constrained. We hypothesise that other proteins would affect the nucleoplasmic solute to act as global “accelerants” or “decelerants” of cellular function and enzymatic activity. We aim to develop innovative methods to understand how cells control protein diffusivity and ultimately develop high-throughput technologies to screen for compounds that target protein kinetics therapeutically.
Building a collaborative and diverse research team
Our goal is to foster an inclusive environment where researchers from various disciplines can work together, bringing their unique perspectives to tackle complex problems. This collaborative approach will drive us to explore a wide range of scientific questions and discoveries.
Publication
- CX-5461 Preferentially Induces Top2α-Dependent DNA Breaks at Ribosomal DNA Loci.
Cameron DP*, Sornkom J*, Alsahafi S, Drygin D, Poortinga G, McArthur GA, Hein N, Hannan RD, Panov KI.
Biomedicines. 12, 1514 (2024).
- Coinhibition of topoisomerase 1 and BRD4-mediated pause release selectively kills pancreatic cancer via readthrough transcription.
Cameron DP*, Grosser J*, Ladigan S*, Kuzin V, Iliopoulou E, Wiegard A, …, Hahn SA, Baranello L.
Science Advances. 9, eadg5109 (2023).
- MYC assembles and stimulates topoisomerases 1 and 2 in a “topoisome”.
Das SK*, Kuzin V*, Cameron DP, Sanford S, Jha RK, Nie Z, Rosello MT, …, Levens D, Baranello L.
Molecular Cell. 82, 140-158.e12 (2022).
- Topoisomerase 1 activity during mitotic transcription favors the transition from mitosis to G1.
Wiegard A*, Kuzin V*, Cameron DP*, Grosser J, Ceribelli, Michele, …, Baranello L.
Molecular Cell. 81, 5007-5024.e9 (2021).
- Analysis of Myc Chromatin Binding by Calibrated ChIP-Seq Approach.
Cameron DP*, Kuzin V*, Baranello L.
Methods in Molecular Biology. 2318, 161-185 (2021).
- Deficiency of Polη in Saccharomyces cerevisiae reveals the impact of transcription on damage-induced cohesion.
Wu PS, Grosser J*, Cameron DP*, Baranello L, Ström L.
PLOS Genetics. 17, e1009763 (2021).
- First-in-Human RNA Polymerase I Transcription Inhibitor CX-5461 in Patients with Advanced Hematological Cancers: Results of a Phase I Dose Escalation Study.
Khot A*, Brajanovski N*, Cameron DP, Hein N, Maclachlan KH, Sanij E, ..., Harrison SJ.
Cancer Discovery. 9, 1036-1049 (2019).
- Inhibition of Pol I transcription treats murine and human AML by targeting the leukemia-initiating cell population.
Hein N, Cameron DP, Hannan KM, Nguyen NN, Fong CY, Sornkom J, …, Hannan RD
Blood. 129, 2882-2895 (2017).
- Selective inhibition of RNA polymerase I transcription as a potential approach to treat African trypanosomiasis
Kerry LE, Pegg EE, Cameron DP, Budzak J, Poortinga G, Hannan K, Hannan RD, Rudenko G.
PLOS Neglected Tropical Diseases. 11, e0005432 (2017).
Donald Cameron
Associate Professor
Ph.D.
Andrej Paluda
Project Research Associate
Ph.D.