Theme
Our laboratory focuses on “translation”, the process by which mRNAs make proteins. Aiming to understand the hidden secrets of this process, we observe translational regulation in situ at single-cell and single-mRNA resolution. Additionally, we strive to manipulate translation at the single-cell and single-mRNA levels.
About Research
mRNA translation at single-cell and single-molecule resolution
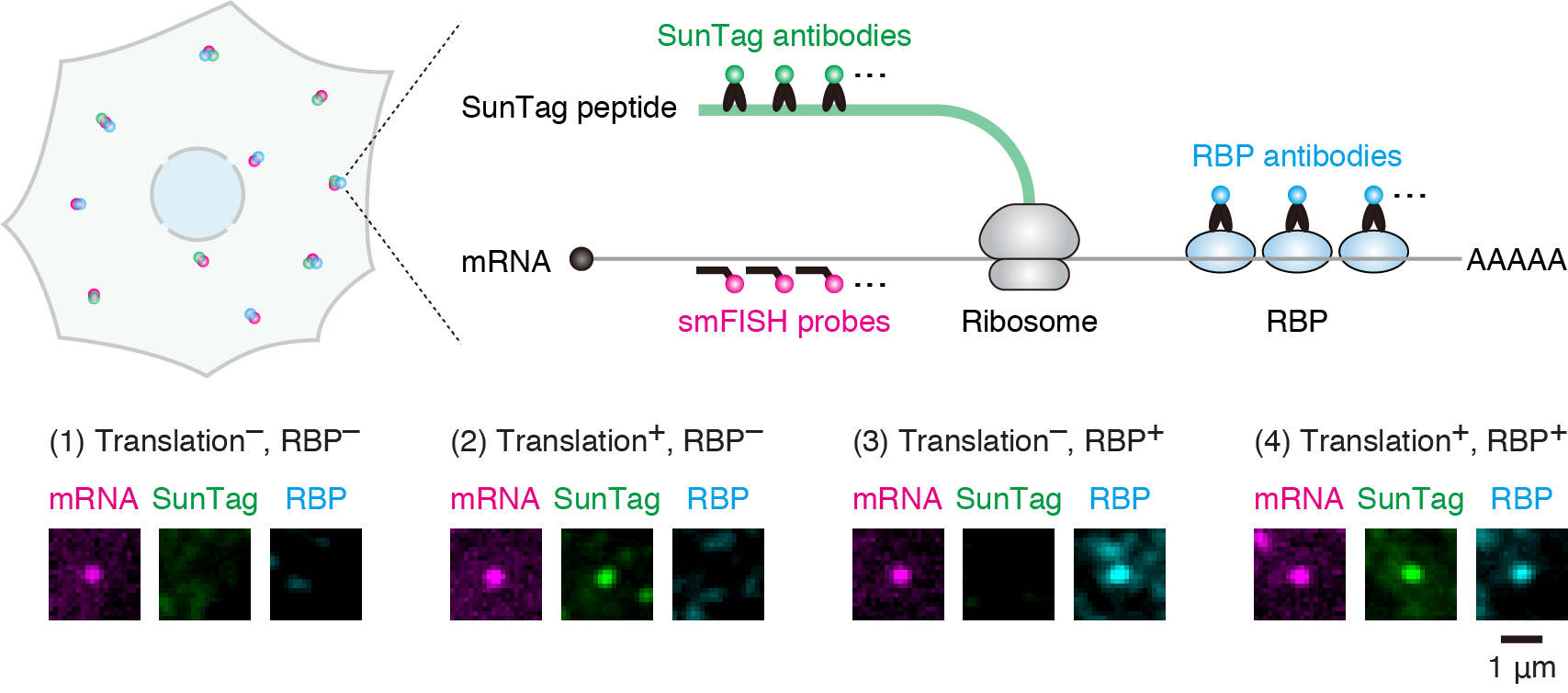
The human genome contains around 20,000 protein-coding genes. For these genes to be expressed, their DNA sequences are transcribed into mRNAs, which are then translated into proteins. Our laboratory focuses on the latter part of gene expression known as “translation”.
As translation is a fundamental process of life, it has been extensively studied for over half a century. However, an issue is that most of the past research on translation has predominantly relied on biochemical methods. While biochemical methods are powerful for addressing molecular mechanisms of “how” translation is regulated, experiments conducted in test tubes fail to obtain spatiotemporal information inside cells. As such, it remains mostly unknown “when” and “where” translation is regulated in cells. Furthermore, traditional biochemical approaches, which analyze a bulk collection of cells and mRNAs collectively, have overlooked the behavior of individual cells and mRNAs for several decades.
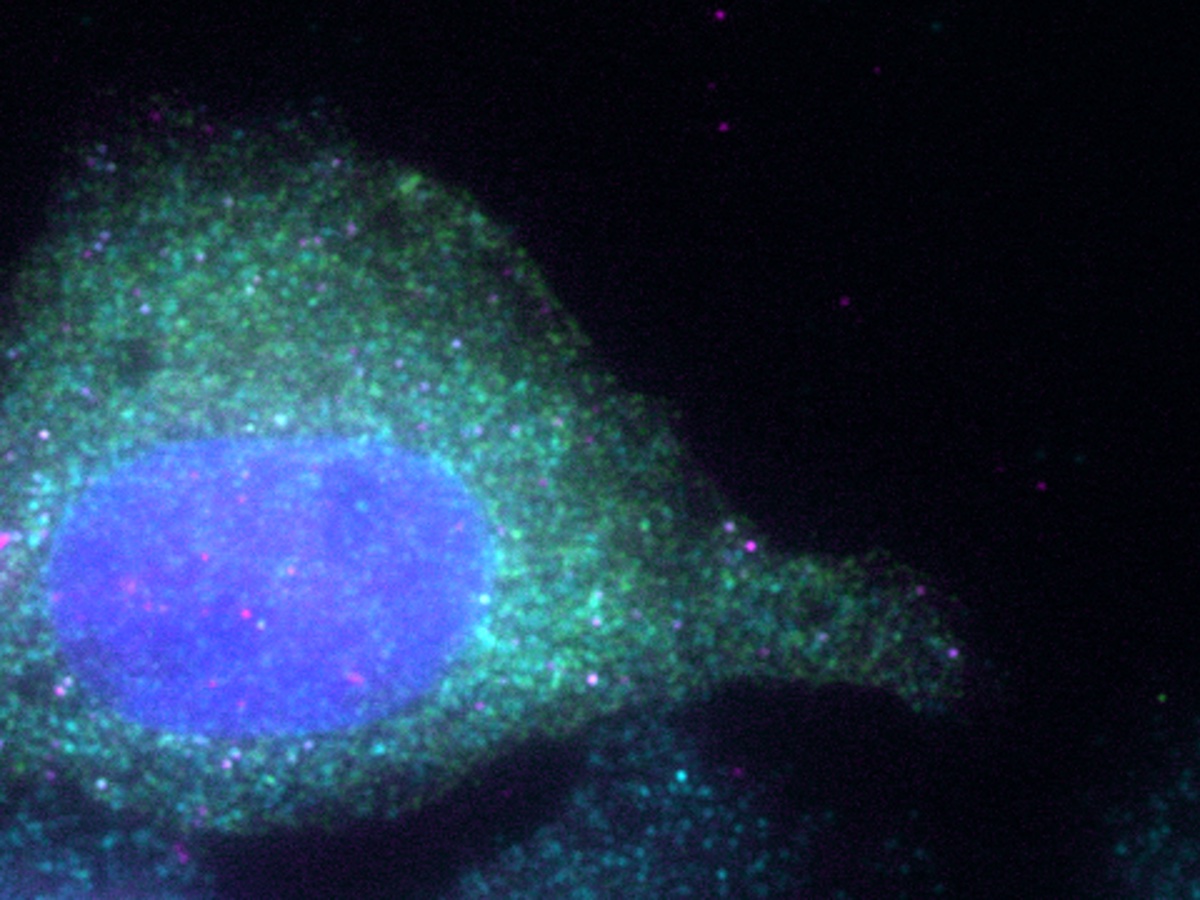
To explore the unknown of translation, we have developed a novel method to image translational regulation as it is at single-cell and single-mRNA resolution. Leveraging this method, our laboratory observes translational regulation of individual cells and mRNAs in situ to understand the hidden secrets of this process – the spatiotemporal aspects of translation, as well as the behavior of individual cells and mRNAs. Additionally, we strive to develop a novel technique to manipulate translation at the single-cell and single-mRNA levels.
Publication
- Single-molecule imaging of microRNA-mediated gene silencing in cells.
Hotaka Kobayashi, Robert H Singer.
Nature Communications. 13(1):1435. 2022
- Identification of an AGO (Argonaute) protein as a prey of TER94/VCP.
Hotaka Kobayashi, Yukihide Tomari.
Autophagy. 16(1):190-192. 2020
- VCP machinery mediates autophagic degradation of empty Argonaute.
Hotaka Kobayashi, Keisuke Shoji, Kaori Kiyokawa, Lumi Negishi, Yukihide Tomari.
Cell Reports. 28(5):1144-1153. 2019
- Iruka eliminates dysfunctional Argonaute by selective ubiquitination of its empty state.
Hotaka Kobayashi, Keisuke Shoji, Kaori Kiyokawa, Lumi Negishi, Yukihide Tomari.
Molecular Cell. 73(1):119-129. 2019
- RISC assembly: Coordination between small RNAs and Argonaute proteins.
Hotaka Kobayashi, Yukihide Tomari.
Biochimica et Biophysica Acta. 1859(1):71-81. 2016
- Measurement of Rab35 activity with the GTP-Rab35 trapper RBD35.
Hotaka Kobayashi, Kan Etoh, Soujiro Marubashi, Norihiko Ohbayashi, Mitsunori Fukuda.
Methods in Molecular Biology. 1298:207-216. 2015
- Rab35 promotes the recruitment of Rab8, Rab13, and Rab36 to recycling endosomes through MICAL-L1 during neurite outgrowth.
Hotaka Kobayashi, Kan Etoh, Norihiko Ohbayashi, Mitsunori Fukuda.
Biology Open. 3(9):803-814. 2014
- Arf6, Rab11, and transferrin receptor define distinct populations of recycling endosomes.
Hotaka Kobayashi, Mitsunori Fukuda.
Communicative & Integrative Biology. 5:e25036. 2013
- Rab35 establishes the EHD1-association site by coordinating two distinct effectors during neurite outgrowth.
Hotaka Kobayashi, Mitsunori Fukuda.
Journal of Cell Science. 126(11):2424-2435. 2013
- Rab35 regulates Arf6 activity through centaurin-β2 (ACAP2) during neurite outgrowth.
Hotaka Kobayashi, Mitsunori Fukuda.
Journal of Cell Science. 125(9):2235-2243. 2012
Hotaka Kobayashi
Visiting Associate Professor
Ph.D.