Theme
RNAs are modified post-transcriptionally to acquire their intrinsic functions. We are investigating functional roles of RNA modifications in various steps of gene expression.
About Research
RNA modifications associated with various biological functions
RNA molecules are frequently modified post-transcriptionally and these modifications are required for proper RNA functions. To date, over 160 different types of chemical modifications have been identified in various RNAs across all domains of life. Recent studies stress the importance of RNA modifications as regulatory elements in gene expression, and the process based on RNA modification is also referred to as “epitranscriptome”. We developed our platform technologies for isolation of individual RNA molecules and highly sensitive analysis of RNA modifications using mass spectrometry (RNA-MS). These platform technilogies enable us to discover novel RNA modifications and RNA-modifying enzymes. Moreover, we found the first instance of human disease caused by lack of RNA modification, and proposed “RNA modopathy” as a new category of human disease. We are tackling to elucidate various biological phenomena associated with RNA functions.
Publication
- Akichika, S.+, Hirano, S.+, Shichino, Y., Sugita, A., Suzuki, T., Nishimasu, H., Ishitani, R., Sugita, A., Hirose, Y., Iwasaki, S., *Nureki, O. and *Suzuki, T. + equal contribution
Cap-specific terminal N6-methylation of RNA by an RNA polymerase II-associated methyltransferase
Science, in press (2018)
- Taniguchi, T., Miyauchi, K., Sakaguchi, Y., Yamashita, S., Soma, A., Tomita, K. and *Suzuki, T.
Acetate-dependent tRNA acetylation required for decoding fidelity in protein synthesis
Nature Chem Biol., 14, 1010-1020 (2018)
- Lin, H., Miyauchi, K., Harada, T., Okita, R., Takeshita, E., Komaki, H., Yagasaki, H., Fujioka, K., Goto, Y., Yanaka, K., Nakagawa, S., Sakaguchi, Y. and *Suzuki, T.
CO2-sensitive tRNA modification associated with human mitochondrial disease
Nature Commun., 14, 9(1):1875 (2018)
- Nagao, A., Ohara, M., Miyauchi, K., Yokobori, S., Yamagishi, A., Watanabe, K. and *Suzuki, T.
Hydroxylation of a conserved tRNA modification establishes non-universal genetic code in echinoderm mitochondria
Nature Struct Mol Biol., 24, 778-782 (2017)
- Ohira, T. and *Suzuki, T.
Precursors of tRNAs are stabilized by methylguanosine cap structures
Nature Chem Biol., 12, 648-655 (2016)
- Nakano, S.+, Suzuki, T. +, Kawarada, L., Iwata, H., Asano, K. and *Suzuki, T.
NSUN3 methylase initiates 5-formylcytidine biogenesis in human mitochondrial tRNAMet
Nature Chem Biol., 12, 546-551 (2016) + equal contribution
- *Frye, M., *Jaffrey, S., *Pan, T., *Rechavi, G. and *Suzuki, T.
RNA modifications: what have we learned and where are we headed?
Nature Rev Genet., 17, 365-372 (2016)
- Miyauchi, K., Kimura, S. and *Suzuki, T.
A cyclic form of N6-threonylcarbamoyladenosine as a widely distributed tRNA hypermodification
Nature Chem Biol., 9, 105-111 (2013)
- Terasaka, N., Kimura, S., Osawa, T., Numata, T. and *Suzuki, T.
Biogenesis of 2-agmatinylcytidine catalyzed by the dual protein and RNA kinase TiaS
Nature Struct Mol Biol., 18, 1268-1274 (2011)
- Sakurai, M., Yano, T., Kawabata, H., Ueda, H. and *Suzuki, T.
Inosine cyanoethylation identifies A-to-I RNA editing sites in the human transcriptome
Nature Chem Biol., 6, 733-740 (2010)
Tsutomu Suzuki
Professor
Ph. D.
Graduate School of Engineering

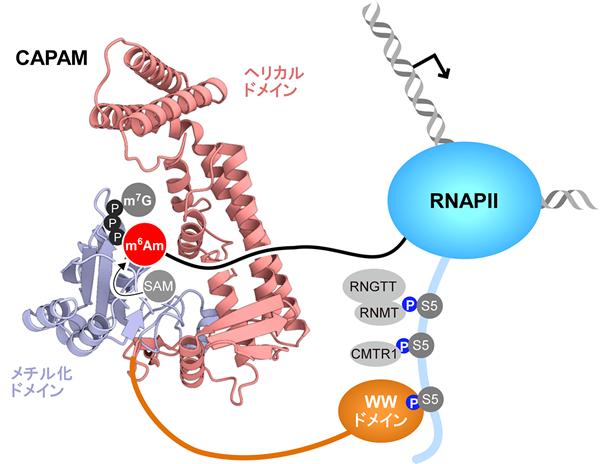

CAPAM is recruited to the early elongation stage of RNA polymerase II via specific interaction between the WW domain and Ser5-phosphorylated CTD. The m7G cap methyltransferase (RNMT) complexed with the capping enzyme (RNGTT) and 2′-O-methyltransferase (CMTR1) are also recruited to this complex, indicating a hierarchical formation of m7Gpppm6Am—pppA, GpppA, m7GpppA, m7GpppAm, and m7Gpppm6Am