Theme
We use DNA origami and DNA circuits and switches to elucidate the biological importance of protein microenvironments on the order of 100nm at the cell surface. We also develop sequencing-based readout methods for spatial biology from the nanoscale to tissue level as well as digital DNA data storage techniques.
About Research
Deciphering cellular interactions on the nanoscale using molecular precision DNA devices
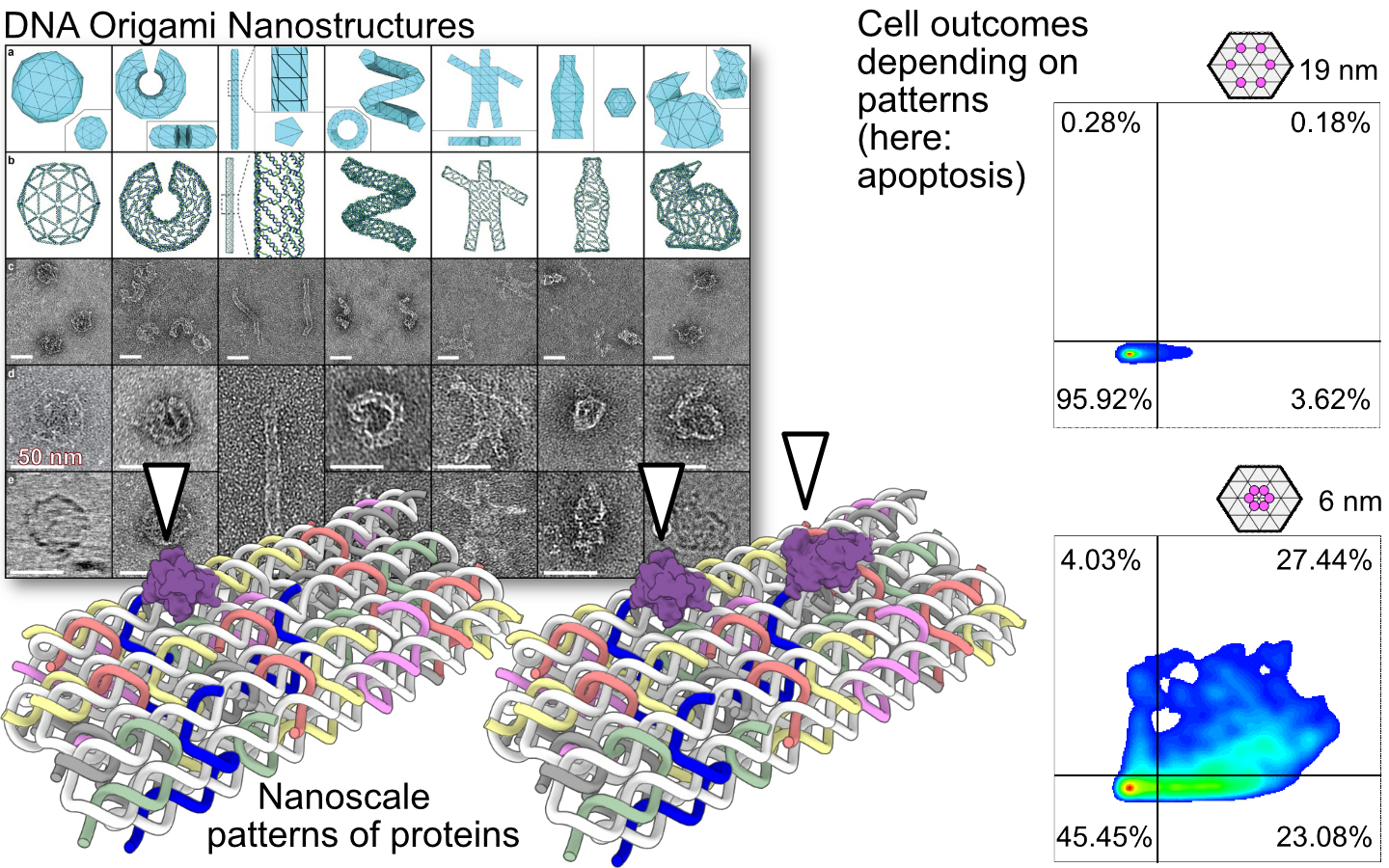
What I cannot create, I cannot understand. This famous quote from American physicist Richard Feynman is a good summation of how we approach solving biological problems in our lab. We believe that in order to understand biology at the nanoscale, we need to be able to manipulate biology at that scale. This is molecular design. We use DNA origami to create assemblies of proteins, peptides and small molecules in nanoscale patterns with molecular precision. This approach allow us to learn how cells communicate using patterns of proteins at the cell membrane in ways that would be impossible to do with traditional methods. Acquiring this type of knowledge of membrane pattern-induced signaling has for example allowed us to design a nano-robotic device that induces apoptosis in tumors, but not elsewhere.
We are also interested in developing new techniques for spatial biology, from the nanoscale and up and how information can be stored and transmitted via DNA. Using DNA in molecular reaction networks, we aim to store and read out neighbor-neighbor interactions. This allows us to read out biological and spatial information at once, from sequencing data alone, without microscopy. The ability to store information in DNA can of course also be used for long term data storage and along the same lines we see synergies with storing spatial information in biological experiments with digital DNA data storage.
Overall, using molecular design and programming we create technique and methods that will help us, and others, to answer fundamental problems about molecular signaling.
Publication
- Wang Y, Baars I, Berzina I, Rocamonde-Lago I, Shen B, Yang Y, Lolaico M, Waldvogel J, Smyrlaki I, Zhu K, Harris RA and Högberg B, A DNA Robotic Switch with Regulated Autonomous Display of Cytotoxic Ligand Nanopatterns, Nature Nanotechnology 19, 1366 (2024)
- Yang Y, Rocamonde-Lago I, Shen B, Berzina I, Zipf J and Högberg B, Re-engineered guide RNA enables DNA loops and contacts modulating repression in E. coli, Nucleic Acids Research 52, 9328 (2024)
- Kloosterman A, Baars I and Högberg B, An error correction strategy for image reconstruction by DNA sequencing microscopy. Nature Computational Science 4, 119 (2024)
- Smyrlaki I, Fördős F, Rocamonde-Lago I, Wang Y, Shen B, Lentini A, Luca VC, Reinius B, Teixeira AI and Högberg B, Soluble and multivalent Jag1 DNA origami nanopatterns activate Notch without pulling force, Nature Communications 15, 465 (2024)
- Wang Y, Benson E, Fördős F, Lolaico M, Baars I, Fang T, Teixeira A & Högberg B, DNA Origami Penetration in Cell Spheroid Tissue Models is Enhanced by Wireframe Design, Advanced Materials 33, p. 2008457 (2021)
- Hoffecker I, Yang Y, Bernardinelli G, Orponen P and Högberg B, A computational framework for DNA sequencing microscopy, Proc. Nat. Acad. Sci. 116, p. 19282–7 (2019)
- Shaw A, Hoffecker IT, Smyrlaki I, Rosa J, Grevys A, Bratlie D, Sandlie I, Michaelsen TE, Andersen JT & Högberg B, Binding to Nanopatterned Antigens is Dominated by the Spatial Tolerance of Antibodies, Nature Nanotechnology, 14 p. 184 (2019)
- Benson E, Mohammed A, Gardell J, Masich S, Czeizler E, Orponen P and Högberg B, DNA rendering of polyhedral meshes at the nanoscale, Nature 523 p. 441 (2015)
- Shaw A, Lundin V, Petrova E, Fördős F, Benson E, Al-Amin A, Herland A, Blokzijl A, Högberg B* and Teixeira A* (*equal), Spatial control of membrane receptor function using ligand nanocalipers, Nature Methods, 11 p. 841 (2014)
- Ducani C, Kaul C, Moche M, Shih WM and Högberg B, Enzymatic production of 'monoclonal stoichiometric' single-stranded DNA oligonucleotides, Nature Methods, 10, p. 647 (2013)
Björn Högberg
Visiting Professor
Ph.D.