Theme
The structure and microenvironment in the eukaryotic nucleus play critical roles in gene regulation. To understand the basic mechanism of transcriptional regulation, we quantitatively measure the dynamics of histone and RNA polymerase II modifications in living cells. Posttranslational modifications on histones and RNA polymerase II play a critical role in transcriptional regulation. To understand the mechanism of gene regulation in the nucleus, we track those modifications in living cells.
About Research
Dynamics of transcription and histone modification in living cells
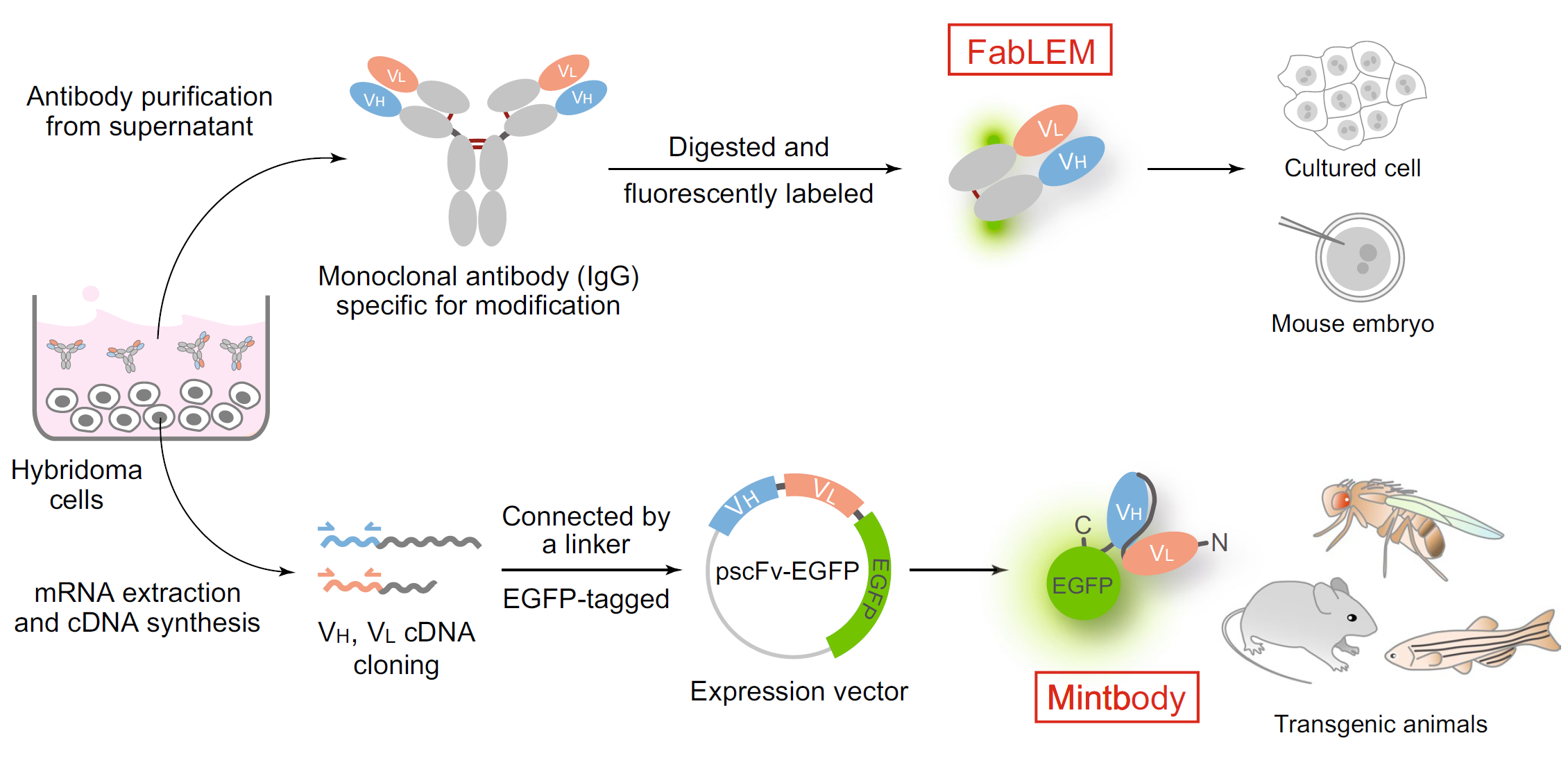
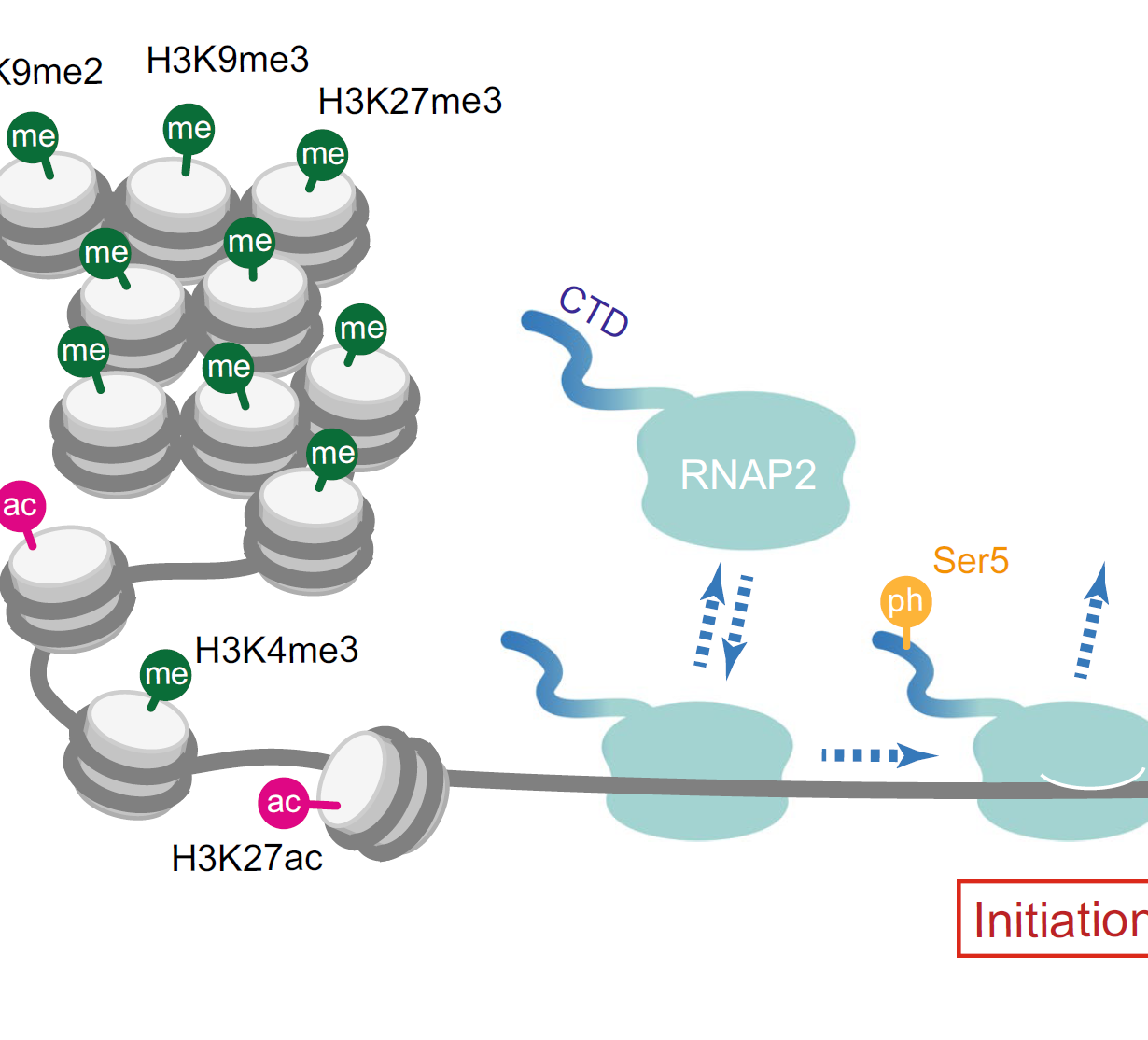
To track the posttranslational modifications of histones and RNA polymerase II, we have generated fluorescent probes for live cell imaging. In particular, genetically encoded modification-specific intracellular antibodies or“ mintbodies” have enabled long-term and in vivo imaging. By measuring the kinetics of transcription activation of a steroid hormone responsive gene array, we revealed the function of histone H3 acetylation to stimulate the transition from the initiation to elongation of RNA polymerase II. We are now tracking the changes in histone modifications during heat-shock response, zygotic genome activation, and X-chromosome inactivation. We are also developing a method for low-input epigenomic profiling applicable to the imaged cells.
Publication
- Sato Y, Stasevich TJ, Kimura H. Visualizing the Dynamics of Inactive X Chromosomes in Living Cells Using Antibody-Based Fluorescent Probes. Methods Mol Biol. 2018;1861:91-102.
- Sato Y, Kujirai T, Arai R, Asakawa H, Ohtsuki C, Horikoshi N, Yamagata K, Ueda J, Nagase T, Haraguchi T, Hiraoka Y, Kimura A, Kurumizaka H, Kimura H. A Genetically Encoded Probe for Live-Cell Imaging of H4K20 Monomethylation. J Mol Biol. 2016 Oct 9;428(20):3885-3902
- Kaimori JY, Maehara K, Hayashi-Takanaka Y, Harada A, Fukuda M, Yamamoto S, Ichimaru N, Umehara T, Yokoyama S, Matsuda R, Ikura T, Nagao K, Obuse C, Nozaki N, Takahara S, Takao T, Ohkawa Y, Kimura H, Isaka Y. Histone H4 lysine 20 acetylation is associated with gene repression in human cells. Sci Rep. 2016, 6: 24318. doi: 10.1038/srep24318
- Hayashi-Takanaka Y, Maehara K, Harada A, Umehara T, Yokoyama S, Obuse C, Ohkawa Y, Nozaki N, Kimura H. Distribution of histone H4 modifications as revealed by a panel of specific monoclonal antibodies. Chromosome Res. 2015, 23(4): 753-66. doi: 10.1007/s10577-015-9486-4
- Kimura H, Yamagata K. Visualization of epigenetic modifications in preimplantation embryos. Methods Mol Biol. 2015, 1222: 127-147. doi: 10.1007/978-1-4939-1594-1_10.
Kimura H, Hayashi-Takanaka Y, Stasevich TJ, Sato Y. Visualizing posttranslational and epigenetic modifications of endogenous proteins in vivo. Histochem Cell Biol. 2015, 144(2): 101-109. doi: 10.1007/s00418-015-1344-0
- Dias JD, Rito T, Torlai Triglia E, Kukalev A, Ferrai C, Chotalia M, Brookes E, Kimura H, Pombo A. Methylation of RNA polymerase II non-consensus Lysine residues marks early transcription in mammalian cells. Elife. 2015, 4. pii: e11215. doi: 10.7554/eLife.11215
- Stasevich TJ, Hayashi-Takanaka Y, Sato Y, Maehara K, Ohkawa Y, Sakata-Sogawa K, Tokunaga M, Nagase T, Nozaki N, McNally JG, Kimura H. Regulation of RNA polymerase II activation by histone acetylation in single living cells. Nature. 2014, 516(7530): 272-275. doi: 10.1038/nature13714
- Stasevich TJ, Sato Y, Nozaki N, Kimura H. Quantifying histone and RNA polymerase II post-translational modification dynamics in mother and daughter cells. Methods. 2014, 70(2-3): 77-88. doi: 10.1016/j.ymeth.2014.08.002
- Hayashi-Takanaka Y, Stasevich TJ, Kurumizaka H, Nozaki N, Kimura H. Evaluation of chemical fluorescent dyes as a protein conjugation partner for live cell imaging. PLoS One. 2014, 9(9):e106271. doi: 10.1371/journal.pone.0106271
Hiroshi Kimura
Professor
Ph. D.