Theme
Our lab uses quantitative live imaging methods to investigate how cis -regulatory DNA elements control transcription dynamics in development.
About Research
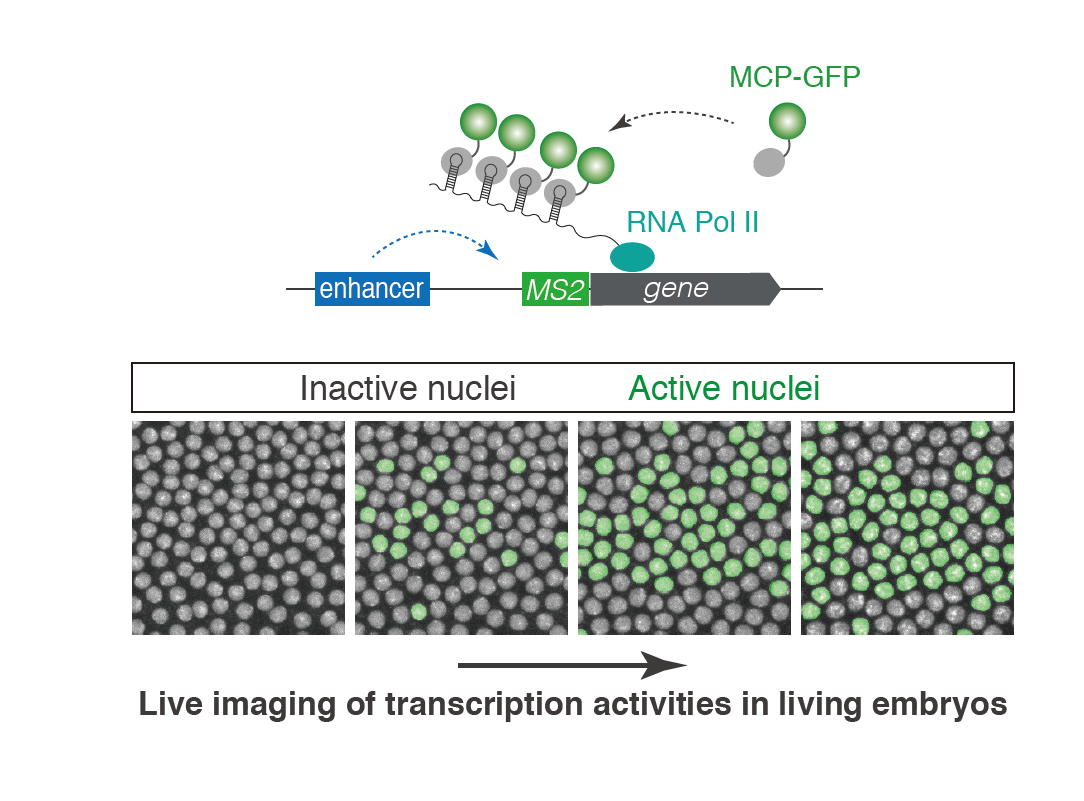
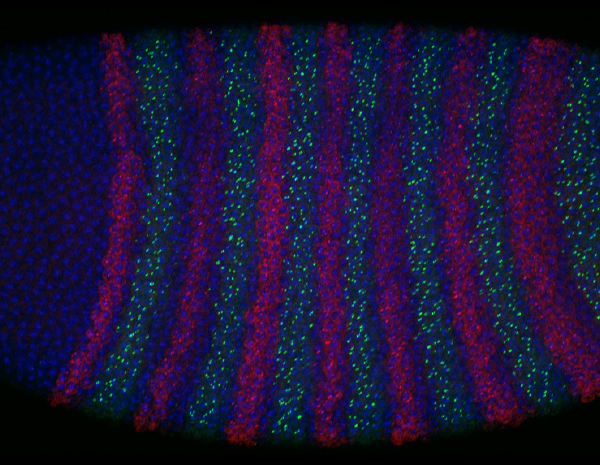
Visualizing enhancer dynamics in living embryos
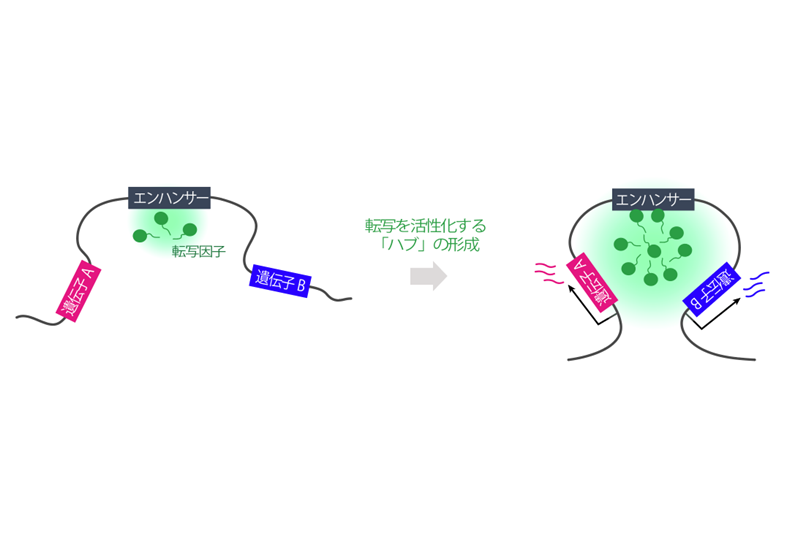
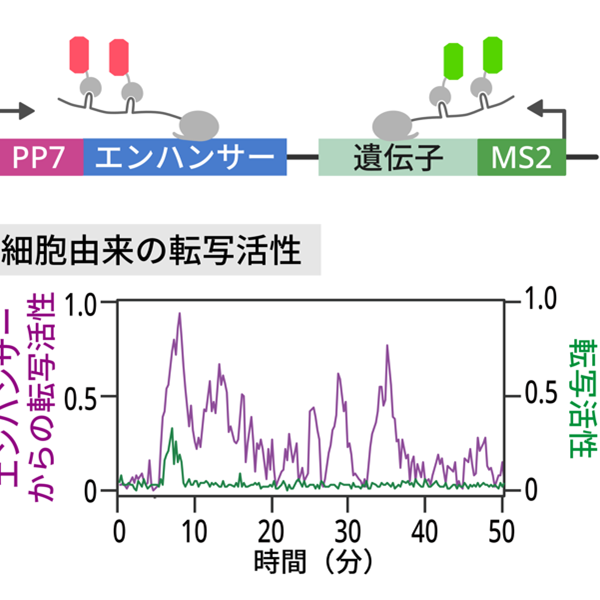
Enhancers are regulatory DNAs that control spatial and temporal dynamics of gene expression in development. Through interaction with sequence-specific transcription factors and co-activators, enhancers modulate when and where genes should be transcribed in response to intringic and extrinsic signals. Recent whole-genome studies estimated that the human genome contains approximately 900,000 enhancers, suggesting that a typical human gene is regulated by 〜40-50 enhancers. Mutation in enhancers can cause dysregulation of transcriptional program, which in turn leads to human diseases such as cancer. Growing number of evidences suggest that diversification of enhancer function is a major source of phenotypic polymorphism among population. While enhancer’s biological importance is becoming increasingly clear in recent years, very little is known about the basic mechanism of enhancer function in the control of gene expression. Especially, the nature of enhancer-promoter communication and its dynamics remain as an outstanding mystery in genome biology. By using a wide-range of experimental approaches such as live-imaging, genome-editing, super-resolution microscopy and whole-genome sequencing methods, we aim to elucidate fundamentally new dimension of the molecular mechanism underlying transcriptional control in development.
Publication
- Enhancer dynamics: Unraveling the mechanism of transcriptional bursting.
Fukaya T.
Science Advances. 9(31), eadj3366. (2023)
- Functional coordination between transcription factor clustering and gene activity.
Kawasaki K, Fukaya T.
Molecular Cell. 83(10):1605-1622. (2023)
- Dynamic interplay between non-coding enhancer transcription and gene activity in development.
Hamamoto K, Umemura Y, Makino S, Fukaya T.
Nature Communications. 14(1):826. (2023)
- Mod(mdg4) variants repress telomeric retrotransposon HeT-A by blocking subtelomeric enhancers.
Takeuchi C, Yokoshi M, Kondo S, Shibuya A, Saito K, Fukaya T, Siomi H, Iwasaki Y.W.
Nucleic Acids Research. 50(20):11580-11599. (2022)
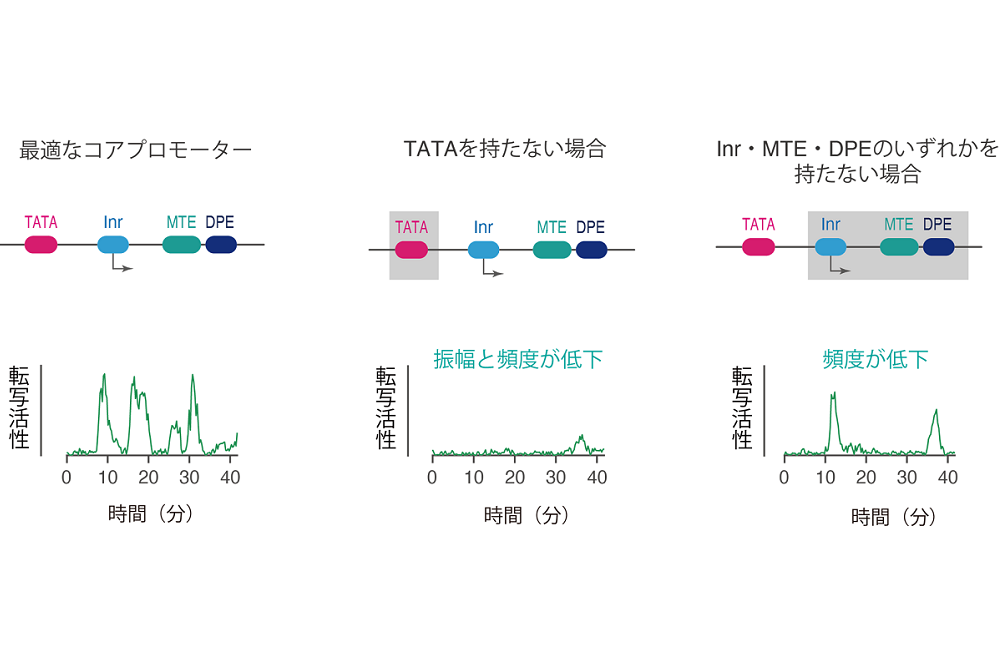
- Dynamic modulation of enhancer responsiveness by core promoter elements in living Drosophila embryos.
Yokoshi M, Kawasaki K (equal contribution), Cambón M, Fukaya T.
Nucleic Acids Research. 50(1):92-107. (2022)
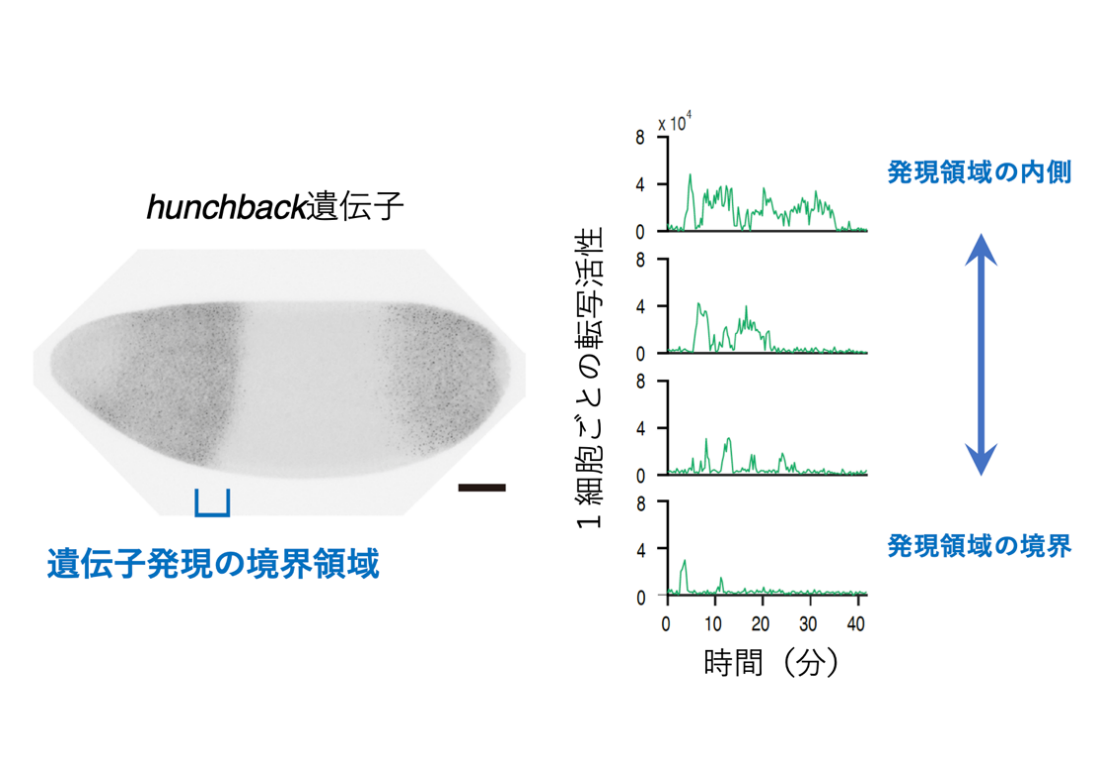
- Dynamic regulation of anterior-posterior patterning genes in living Drosophila embryos.
Fukaya T.
Current Biology. 31(10):2227-2236. (2021)
- Tissue-scale mechanical coupling reduces morphogenetic noise to ensure precision during epithelial folding.
Eritano A.S, Bromley C.L, Bolea Albero A, Schütz L, Wen F.-L, Takeda M, Fukaya T, Sami M.M, Shibata T, Lemke S, and Wang Y.-C.
Developmental Cell. 53(2):212-228. (2020)
- Visualizing the role of boundary elements in enhancer-promoter communication.
Yokoshi M, Segawa K, Fukaya T.
Molecular Cell. 78(2):224-235. (2020)
- Visualization of transvection in living Drosophila embryos
Lim B, Heist T, Levine M, Fukaya T.
Molecular Cell. 70 (2), 287-296. (2018)
- Enhancer control of transcriptional bursting.
Fukaya T, Lim B, Levine M.
Cell. 166 (2), 358-368. (2016)
Takashi Fukaya
Professor
Ph.D.
Graduate School of Arts and Sciences
Shiho Makino
Research Associate
Ph.D.
Raku Saito
Project Research Associate
Ph.D.
Koji Kawasaki
Project Research Associate
Ph.D.