Theme
We design the self-assembly of DNA and RNA, leveraging existing structural motifs and enzymatic reactions to create nanoscale machines, sensors, and other advanced molecular architectures. Our aim is to advance synthetic biology, biophysics, and beyond by developing new bio-tools that deepen our understanding of fundamental biological phenomena.
About Research
Tailoring DNA and RNA into groundbreaking molecular tools that drive next-generation bioengineering
DNA and RNA are not merely carriers of genetic information but also extraordinary materials that can be precisely engineered for self-assembly and specific interactions. Our research focuses on “nucleic acid nanotechnology,” exploiting these features to design nanoscale machines, sensors, and other advanced molecular robots.
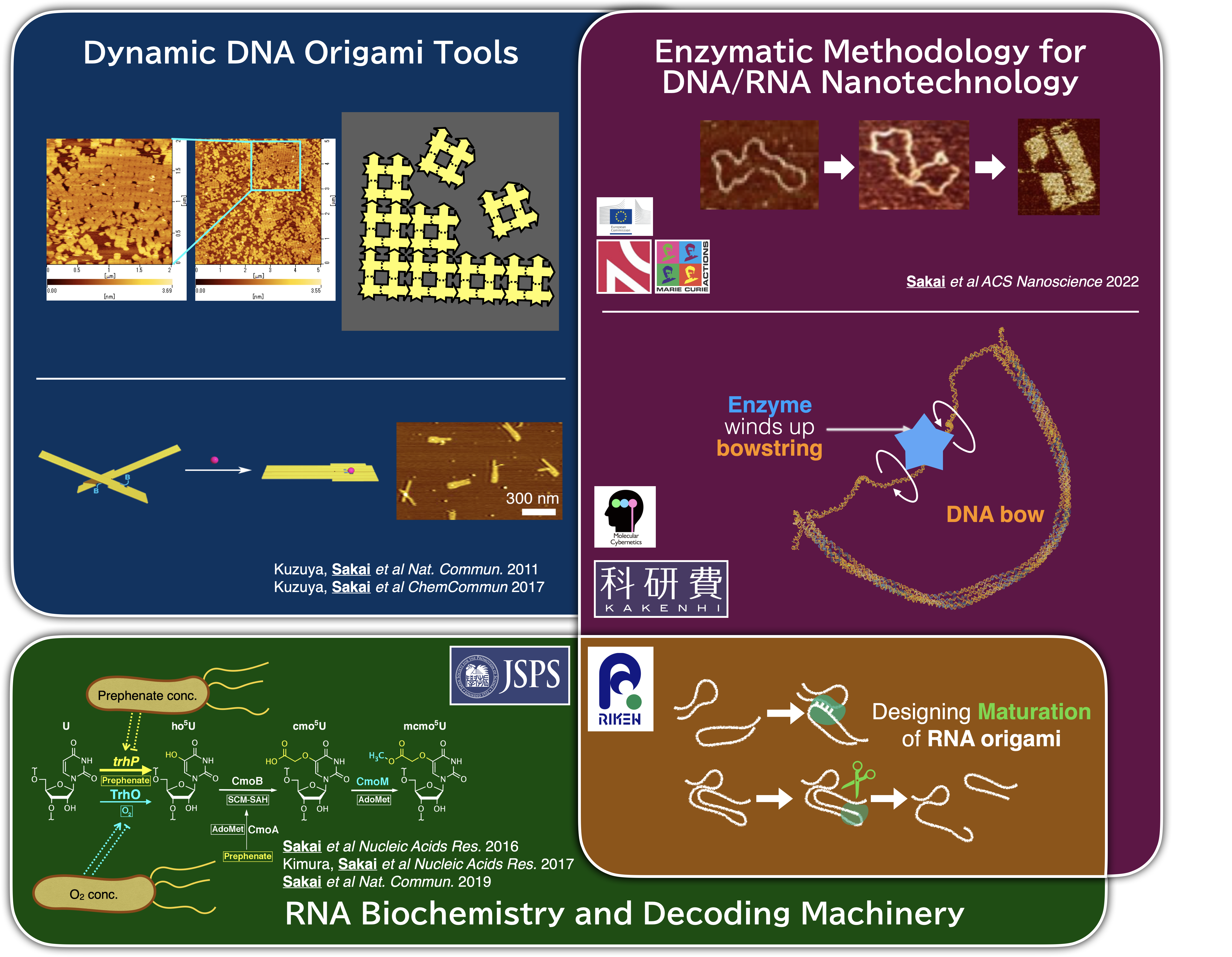
We have developed modular DNA-based sensors, nanoscale capsules isolating internal cargo, and DNA origami platforms for arranging molecular components over large areas. By leveraging enzymes refined through evolution, we integrated DNA topoisomerases to catalyse the catenation of long DNA strands, enabling topologically interlinked DNA origami. Currently, we are exploring another DNA topoisomerase as a power source for nanodevices and investigating sophisticated RNA origami in vivo using abundant ribosome assembly factors.
Our collaborations span synthetic biology and biophysics, including joint projects at RIKEN, Institute of Science Tokyo, Tokyo University of Agriculture and Technology, and Durham University (UK). To handle large sets of oligo DNA and their mixing patterns, we implement specialised database software and automated instrumentation, enhancing both effectient collaborative studies and large-scale molecular system development. Ultimately, we aim to create versatile and scalable molecular tools that deepen our understanding of fundamental biological questions. This research is supported by the “UTokyo-KI LINK” programme between the University of Tokyo and Karolinska Institutet, with our group is located in Professor Björn Högberg’s laboratory at Karolinska Institutet.
Publication
- Gerrit D. Wilkens, Piotr Stepien, Yusuke Sakai, Sirajul Md. Islam, Jonathan G Heddle*. “A DNA origami bubble blower for liposome production.” ACS Omega, 9, 43, 43609-43615 (2024) DOI: 10.1021/acsomega.4c05297
- Yusuke Sakai,† Gerrit D. Wilkens,† Karol Wolski, Szczepan Zapotoczny, Jonathan G Heddle*. “TOPOGAMI: Topologically linked DNA origami.” ACS Nanoscience Au, 2, 1, 57-63 (2022) DOI: 10.1021/acsnanoscienceau.1c00027 († equal contribution)
- Yusuke Sakai,† Satoshi Kimura,† Tsutomu Suzuki. “Dual pathways of tRNA hydroxylation ensure efficient translation by expanding decoding capability.” Nature Communications, 10, 2858 (2019) DOI: 10.1038/s41467-019-10750-8 († equal contribution)
- Yusuke Sakai, Md. Sirajul Islam, Martyna Adamiak, Simon Chi-Chin Shiu, Julian Alexander Tanner, Jonathan Gardiner Heddle,* “DNA Aptamers for Functionalisation of DNA Origami Nanostructures.” Genes, 9, 571 (2018) DOI: 10.3390/genes9120571 (Review)
- Simon Chi-Chin Shiu, Andrew B Kinghorn, Yusuke Sakai, Yee-Wai Cheung, Jonathan G Heddle, and Julian A Tanner,* “The Three S's for Aptamer-Mediated Control of DNA Nanostructure Dynamics: Shape, Self-Complementarity, and Spatial Flexibility.” ChemBioChem, 19, 18, 1900–1906 (2018) DOI: 10.1002/cbic.201800308 (Review)
- Satoshi Kimura, Yusuke Sakai, Kenta Ishiguro, Tsutomu Suzuki,* “Biogenesis and iron-dependency of ribosomal RNA hydroxylation.” Nucleic Acids Research, 45, 22, 1–14 (2017) DOI: 10.1093/nar/gkx969
- Akinori Kuzuya,* Yusuke Sakai, Takahiro Yamazaki, Yan Xu, Yusei Yamanaka, Yuichi Ohya and Makoto Komiyama,* “Allosteric Control of Nanomechanical DNA Origami Pinching Devices for Enhanced Target Binding.” Chemical Communications, 53, 59, 8276–8279 (2017) DOI: 10.1039/C7CC03991C
- Yusuke Sakai, Kenjyo Miyauchi, Satoshi Kimura and Tsutomu Suzuki,* “Biogenesis and growth phase-dependent alteration of 5-methoxycarbonylmethoxyuridine in tRNA anticodons.” Nucleic Acids Research, 44, 509-523 (2016) DOI: 10.1093/nar/gkv1470 (NAR Breakthrough Article selected)
- Takahiro Yamazaki, Yuichiro Aiba, Kohei Yasuda, Yusuke Sakai, Yusei Yamanaka, Akinori Kuzuya,* Yuichi Ohya, and Makoto Komiyama*, “Clear-cut observation of PNA invasion using nano-mechanical DNA origami devices.” Chemical Communications, 48, 11361–11363 (2012) DOI: 10.1039/c2cc36358e
- Akinori Kuzuya,* Yusuke Sakai, Takahiro Yamazaki, Yan Xu, and Makoto Komiyama,* “Nanomechanical DNA origami ‘single-molecule beacons’ directly imaged by atomic force microscopy.” Nature Communications, 2, 449 (2011) DOI: 10.1038/ncomms1452
Yusuke Sakai
Lecturer
Ph. D.