Theme
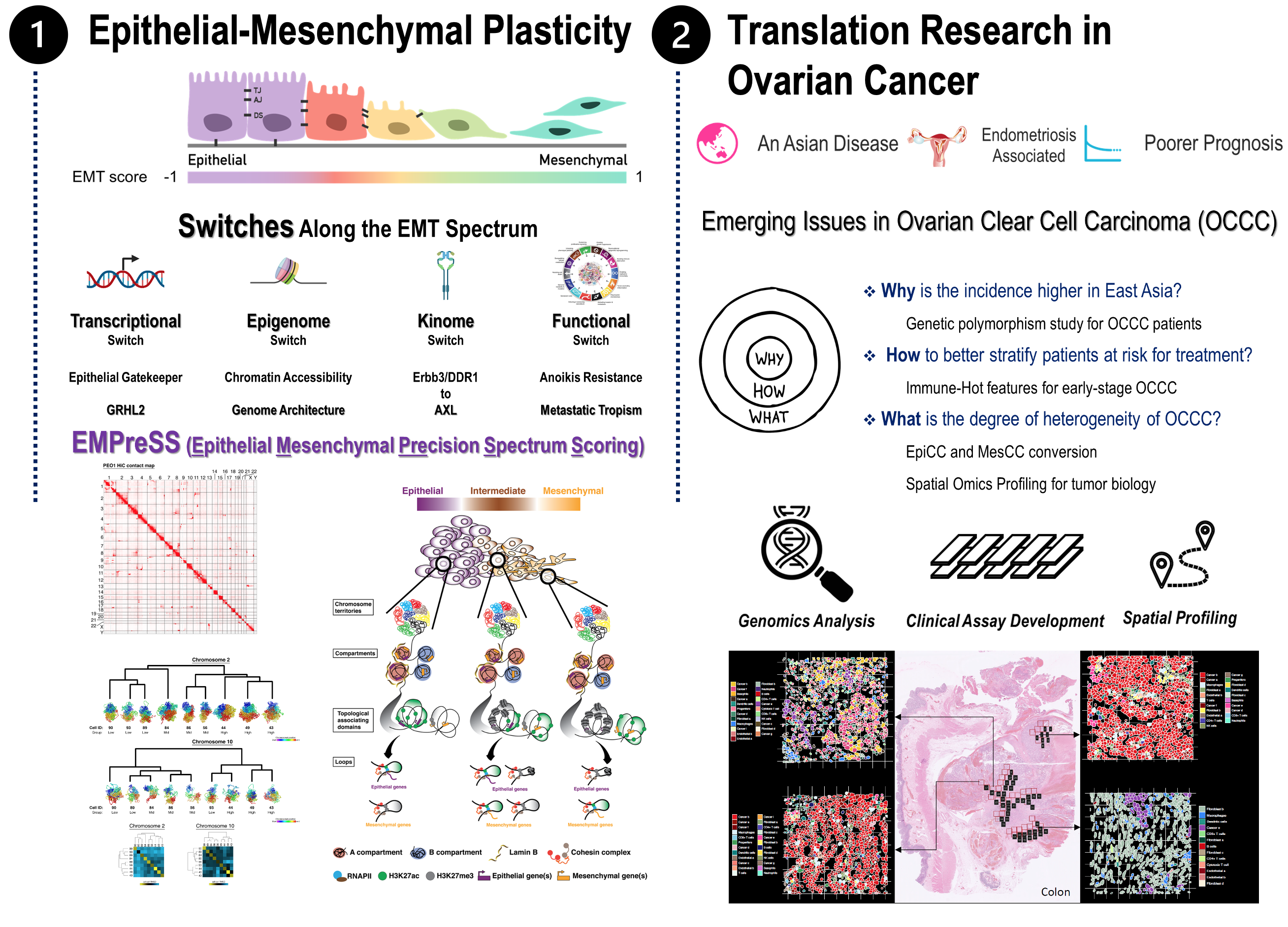
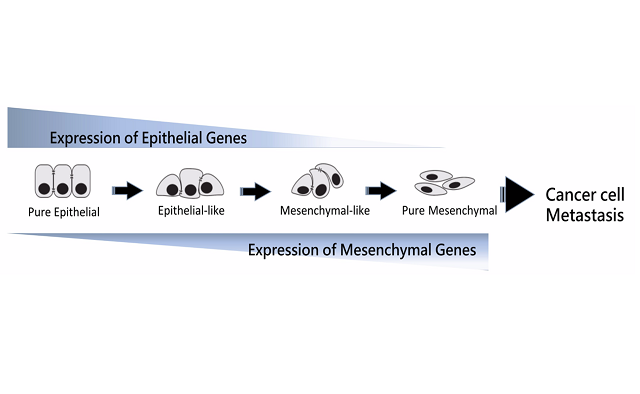
EMPreSS (Epithelial Mesenchymal Precision Spectrum Scoring) assesses epithelial and mesenchymal states beyond binary views. It aims to define epithelial characteristics, precise "mesenchymality," and transitions among states. Recognizing EMT as a spectrum, our lab explores heterogeneity through fluid transition, genome architecture, and spatial context analysis.
About Research
Unlocking Cellular Complexity: Navigate the Epithelial-Mesenchymal Spectrum with EMPreSS
Our EMT investigations redefine the conventional binary model to a continuous spectrum through extensive analysis of over 13,000 cancer gene expression profiles. We establish EMT signatures and scoring mechanisms, elucidating their pivotal role in tumoral heterogeneity. Additionally, our exploration extends to the 3D genome level, shedding light on the intricate regulation of the EMT spectrum.
In the domain of Ovarian Cancer Research, we unveil five distinct molecular subtypes with varying epithelial and mesenchymal characteristics and responses to chemotherapy. Notable pathways such as the non-canonical FZD7-Wnt and AXL signaling pathways are identified, laying the groundwork for personalized treatment strategies. Through initiatives like the Molecular Assessment of Subtype Heterogeneity (MASH), we delve into intra-tumoral heterogeneity, guiding tailored therapeutic interventions.
Moreover, our commitment to Spatial Biology drives us to unravel the complexities of intra-tumoral heterogeneity through advanced spatial genomics techniques. We scrutinize tumor microenvironments and epithelial-mesenchymal plasticity, employing cutting-edge platforms like the nanoString Digital Spatial Profiling (DSP) GeoMx system and the 10X Visium system.
Our multidisciplinary team, encompassing expertise in oncology, pathology, and clinical oncology, collaborates synergistically to advance scientific understanding and translate findings into innovative therapeutic strategies. Our research endeavors hold promise for revolutionizing the management of ovarian cancer and enhancing comprehension of spatial aspects of cancer biology.
Publication
- Huang RY, Huang KJ, Chen KC, Hsiao SM, Tan TZ, Wu CJ, Hsu C, Chang WC, Pan CY, Sheu BC, Wei LH. (2023) Immune-Hot tumor features associated with recurrence in early-stage ovarian clear cell carcinoma. Int J Cancer. 152(10):2174-2185.
- Yu SC, Chen KC, Huang RY. (2023) Nodal reactive proliferation of monocytoid B-cells may represent atypical memory B-cells. J Microbiol Immunol Infect. 56(4):729-738.
- Yeo XH, Sundararajan V, Wu Z, Phua ZJC, Ho YY, Peh KLE, Chiu YC, Tan TZ, Kappei D, Ho YS, Tan DSP, Tam WL, Huang RY. (2023) The effect of inhibition of receptor tyrosine kinase AXL on DNA damage response in ovarian cancer. Commun Biol. 6(1):660.
- Tai YT, Lin WC, Wang DYT, Ye J, Tan TZ, Wei LH, Huang RYJ. (2023) Spatial profiling of ovarian clear cell carcinoma (OCCC) and identification of immune-hot features. Journal of Clinical Oncology. 41 (16_suppl), e17543-e17543.
- Pang QY, Tan TZ, Sundararajan V, Chiu YC, Chee EYW, Chung VY, Choolani MA, Huang RY. (2022) 3D genome organization in the epithelial-mesenchymal transition spectrum. Genome Biol. May 30;23(1):121.
- PY Chu, APF Koh, J Antony, RYJ Huang. (2022) Applications of the Chick Chorioallantoic Membrane as an Alternative Model for Cancer Studies. Cells Tissues Organs. 211 (2), 222-237
- Heong V, Tan TZ, Miwa M, Ye J, Lim D, Herrington CS, Iida Y, Yano M, Yasuda M, Ngoi NY, Wong SJ, Okamoto A, Gourley C, Hasegawa K, Tan DS, Huang RY. (2021) A multi‐ethnic analysis of immune‐related gene expression signatures in patients with ovarian clear cell carcinoma. Journal of Pathology. 255 (3), 285-295
- Sundararajan V, Tan M, Zea Tan T, Pang QY, Ye J, Chung VY, Huang RY. (2020) SNAI1-Driven Sequential EMT Changes Attributed by Selective Chromatin Enrichment of RAD21 and GRHL2. Cancers (Basel). May 2;12(5):1140.
- Chung VY, Tan TZ, Ye J, Huang RL, Lai HC, Wollmann H, Guccione E, Huang RY. (2019) The role of GRHL2 and epigenetic remodeling in epithelial–mesenchymal plasticity. Commun Biol. Jul 24;2:272.
- Tan TZ, Ye J, Yee CV, Lim D, Ngoi NYL, Tan DSP, Huang RY. (2019) Analysis of gene expression signatures identifies prognostic and functionally distinct ovarian clear cell carcinoma subtypes. EBioMedicine. Dec;50:203-210
Ruby Yun-Ju Huang
Visiting Professor
Ph. D.