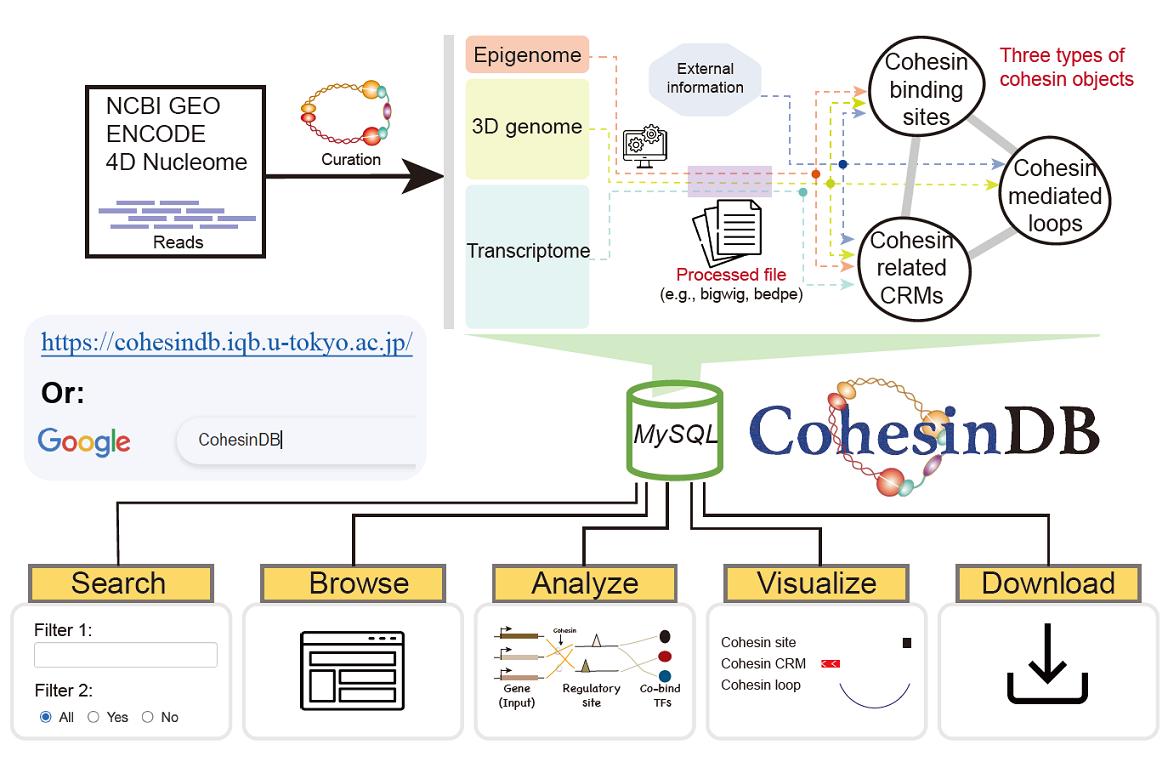
Cohesin is a multifunctional protein responsible for transcriptional regulation and chromatin organization. Even though next-generation sequencing technologies have provided a wealth of information on different aspects of cohesin, the integration and exploration of the resultant massive cohesin datasets are not straightforward. Cohesin can bind to chromatin at tens of thousands of distinct sites, whereas the function of cohesin may varies greatly. Cohesin also extensively mediates cis-regulatory modules (CRMs), genomic regions in which multiple transcription factors bind to regulate gene expression. From a three-dimensional (3D) genomic perspective, cohesin mediates chromatin loops, forming different levels of organization in the nucleus. The big data on cohesin binding, cohesin function, and cohesin-mediated chromatin loops and CRMs are essential not only for the cohesin research field but for all studies of transcriptional regulation and chromosome structure. There is great demand to create a database to summarize cohesin-related information obtained from multiomics datasets.
In this study, Jiankang Wang, a Ph.D. student, and Dr. Ryuichiro Nakato, an associated professor at the Institute for Quantitative Biosciences (IQB), the University of Tokyo, present CohesinDB (https://cohesindb.iqb.u-tokyo.ac.jp/), a comprehensive multiomics cohesin database in human cells. CohesinDB includes 2,043 epigenomics, transcriptomics and 3D genomics datasets from 530 studies involving 176 cell types. By integrating these large-scale data, CohesinDB summarizes three types of ‘cohesin objects’: 751,590 cohesin binding sites, 957,868 cohesin-related chromatin loops and 2,229,500 cohesin-related CRMs. Each cohesin object is annotated with locus, cell type, classification, function, 3D genomics and cis-regulatory information. CohesinDB features a user-friendly interface for browsing, searching, analyzing, visualizing and downloading the desired information. CohesinDB contributes a valuable resource for all researchers studying cohesin, epigenomics, transcriptional regulation and chromatin organization.
Pressrelease
(Go to the IQB Japanese Website)
Journals
Journal:
Nucleic Acids Research
Title: CohesinDB: A comprehensive database for decoding cohesin-related epigenomes, 3D genomes and transcriptomes in human cells
Author: Jiankang Wang and Ryuichiro Nakato*
DOI: 10.1093/nar/gkac795
URL:
https://doi.org/10.1093/nar/gkac795
Contact
Laboratory of Computational Genomics
Associate Professor
Ryuichiro Nakato