研究分野の概略
Accessing the genetic information in the DNA double helix, for example by transcription or DNA replication, generates DNA supercoiling, which in turn can recruit Structural Maintenance of Chromosomes (SMC) protein complexes that initiate folding of chromosomes by DNA loop extrusion. Our research explores how local DNA topology influences the global structure and function of chromosomes.
研究内容の紹介
Untangling the role of DNA supercoiling in chromosome structure and function
The evolutionary conserved Structural Maintenance of Chromosomes (SMC) protein complexes, cohesin, condensin and Smc5/6, are multi-subunit molecular machines that hydrolyze ATP to topologically entrap DNA molecules within their ring-shaped structure, and to fold chromosomes by rapidly extruding DNA loops. Consequently, the SMC complexes are central for the structural organization of chromosomes, and influence most chromosome-based processes, including chromosome segregation, transcription, DNA replication and repair. Cohesin and condensin have been named based on their main functions as cohesin holds sister chromatids together, and condensin compacts chromosomes. The molecular functions of Smc5/6 remain a mystery in the field of chromosome biology, and hence this complex has not been named yet.
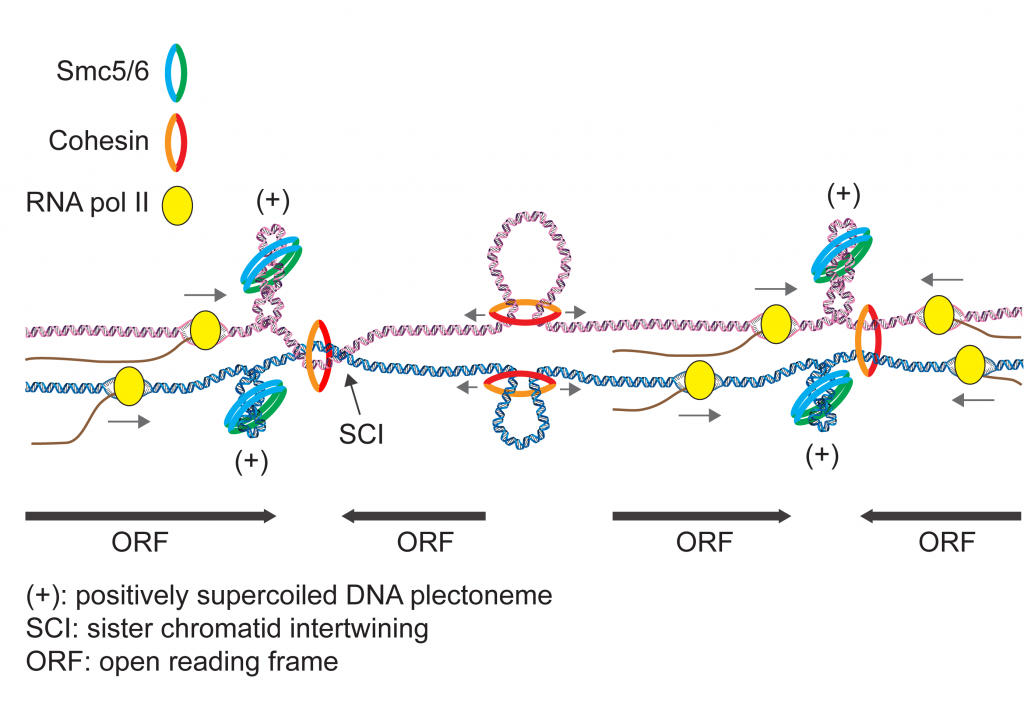
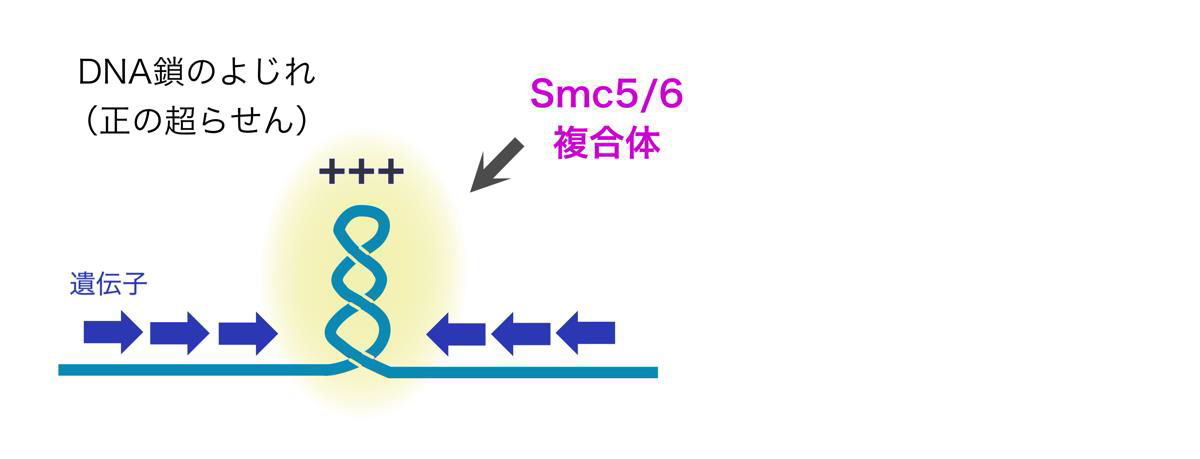
The functions of SMC complexes have been linked to DNA supercoiling, and recently, condensin and Smc5/6 were found to preferentially associate with positively supercoiled DNA in vitro. DNA supercoiling arise in cells when translocating polymerases and helicases unwind the DNA double helix to access the genetic information, resulting in the DNA molecule becoming overtwisted ahead, and undertwisted behind, the translocating machineries. This opens for the intriguing possibility that chromosomal processes that alter local DNA topology, might be closely interlinked with the regulation of chromosome structure through the generation of positive supercoils, which recruit SMC complexes.
Through curiosity-driven basic research combined with state-of-the-art DNA sequencing-based technologies, we aim to elucidate molecular mechanisms of how DNA supercoiling interconnects with global genome organization. Of particular interest, we strive to solve the mystery of how Smc5/6 mechanistically maintains genome stability in cells.
論文一覧
- Jeppsson K*§, Pradhan B, Sutani T, Sakata T, Igarashi MU, Berta DG, Kanno T, Nakato R, Shirahige K, Kim E*, Björkegren C*. Loop-extruding Smc5/6 organizes transcription-induced positive DNA supercoils. Mol Cell. 2024 Mar 7;84(5):867-882.e5. doi: 10.1016/j.molcel.2024.01.005. Epub 2024 Jan 30. (* shared corresponding authors, §lead author).
- Pradhan B*, Kanno T*, Umeda Igarashi M, Dieter Baaske M, Wong JSK, Jeppsson K, Björkegren C*, Kim E*. (2023) The Smc5/6 complex is a DNA loop extruding motor. Nature, 2023 Apr;616(7958):843-848.
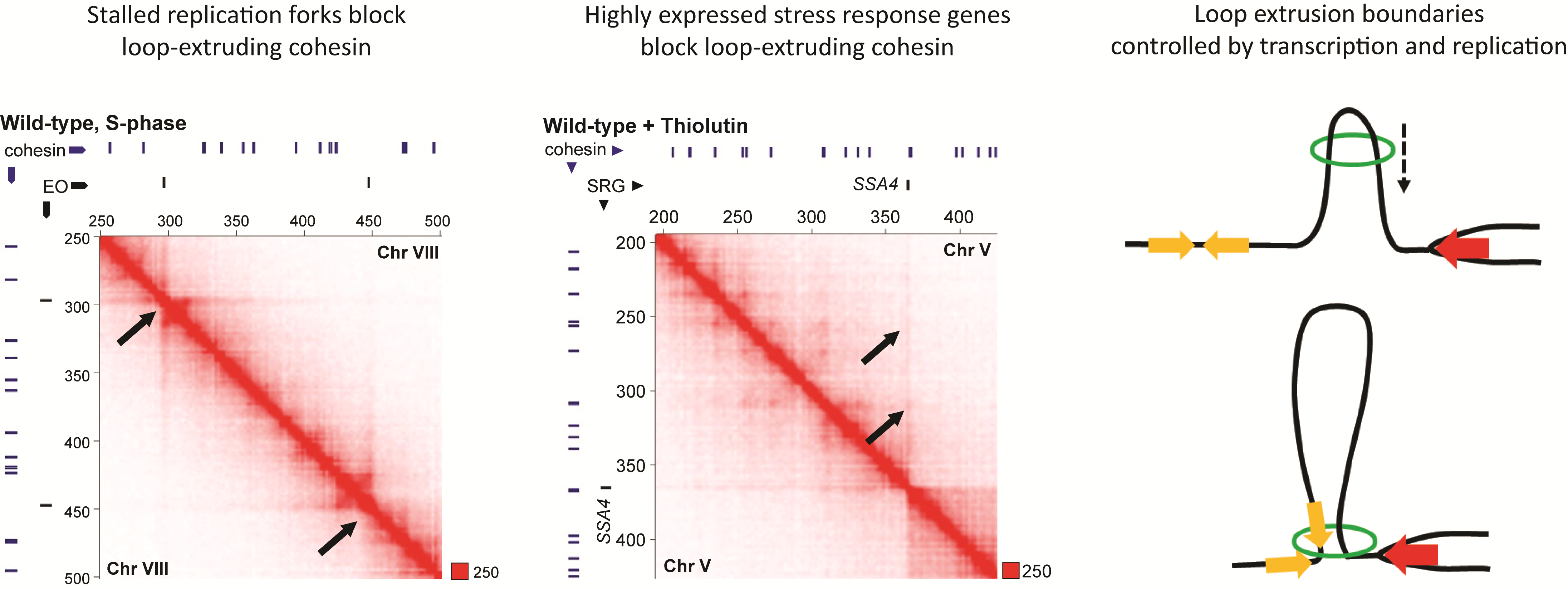
- Jeppsson K*, Sakata T, Nakato R, Milanova S, Shirahige K, Björkegren C*. (2022) Cohesin-dependent chromosome loop extrusion is limited by transcription and stalled replication forks.
Sci Adv. 2022 Jun 10;8(23):eabn7063. , (* shared corresponding authors).
- Jeppsson K, Carlborg KK, Nakato R, Berta DG, Lilienthal I, Kanno T, Lindqvist A, Brink MC, Dantuma NP, Katou Y, Shirahige K, Sjögren C. (2014) The chromosomal association of the Smc5/6 complex depends on cohesion and predicts the level of sister chromatid entanglement. PLoS Genet. 2014 Oct 16;10(10):e1004680.
- Jeppsson K, Kanno T, Shirahige K, Sjögren C. (2014) The maintenance of chromosome structure: positioning and functioning of SMC complexes. Nat Rev Mol Cell Biol. 2014 Aug 22;15(9):601-14.
- Kegel A, Betts-Lindroos H, Kanno T, Jeppsson K, Ström L, Katou Y, Itoh T, Shirahige K, Sjögren C. (2011) Chromosome length influences replication-induced topological stress. Nature 471, 392-396
- Bustard DE, Menolfi D, Jeppsson K, Ball LG, Dewey SC, Shirahige K, Sjogren C, Branzei D, Cobb JA. (2012) During replication stress Non-Smc-Element 5 is required for Smc5/6 complex functionality at stalled forks. J Biol Chem. 2012 Mar 30;287(14):11374-83.
- Burghoorn J, Piasecki BP, Crona F, Phirke P, Jeppsson KE, Swoboda P. (2012) The in vivo dissection of direct RFX-target gene promoters in C. elegans reveals a novel cis-regulatory element, the C-box. Dev Biol. 2012 Aug 15;368(2):415-26.
- Massinen S, Hokkanen ME, Matsson H, Tammimies K, Tapia-Páez I, Dahlström-Heuser V, Kuja-Panula J, Burghoorn J, Jeppsson KE, Swoboda P, Peyrard-Janvid M, Toftgård R, Castrén E, Kere J. (2011) Increased Expression of the Dyslexia Candidate Gene DCDC2 Affects Length and Signaling of Primary Cilia in Neurons. PLoS One. 2011;6(6):e20580.
クリスチャン ヤポセン 講師
Kristian Jeppsson
Ph.D.