It is essential for the survival and growth of living organisms to inherit chromosome (= genetic information) faithfully to next generations. The living organisms have developed the molecular systems by which they replicate chromosomes, repair damages on chromosome, and segregate duplicated chromosomes equally to daughter cells. Chromosome is a platform of life and various so-called chromosomal functions (DNA replication, transcription, recombination, repair, partition, and so on) are integrated into a single molecule of chromosome. Hundreds to thousands of proteins are interacting with chromosomal DNA to perform various chromosomal functions (Fig. 1).
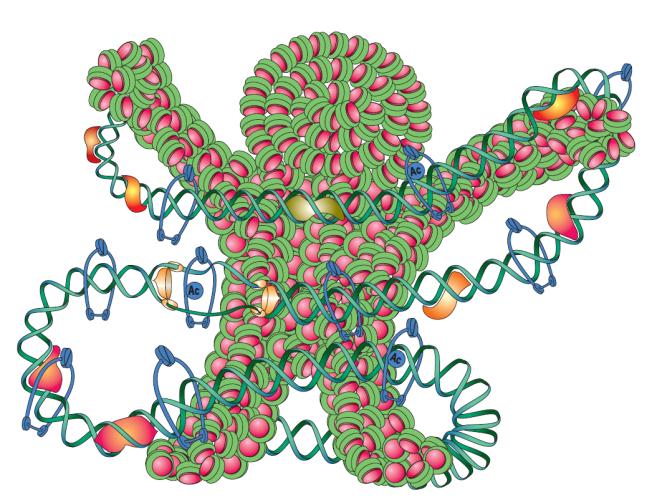
Fig1 Cartoon summarizing concept of chromsome
The methodology called ChIP-chip ( chromatin immunoprecipitation on DNA chip)enables us to study protein-DNA interactions genome wide at several hundreds bp resolution. Our group is one of the pioneers brought this technology in the field of chromosome biology, and the first group to use tiling array (the super high resolution array) for ChIP-chip (nature 2003). Using this technology, we have studied cell cycle dependent or growth condition dependent localization of various chromosomal proteins (Fig2 and Fig3). Protein localization data brings us not only the information of protein complex but also tells us regulatory mechanisms that control protein localization at particular sites on the genome. Especially by comparing various localization profiles of chromosomal proteins, we sometimes see unexpected interplay of chromosome functions. For example, using this technique, we have analyzed how DNA replication checkpoint proteins are integrated into the process of DNA replication, leading us to identification of Mrc1 as a key player for DNA replication checkpoint cascade (nature 2003 and Fig.2). By applying the same technique to the analysis of human chromosome, we have shown that cohesin, a protein complex that is essential for chromosome partitioning, plays also an important role in transcriptional regulation as an insulator protein (nature 2008 and Fig 3).
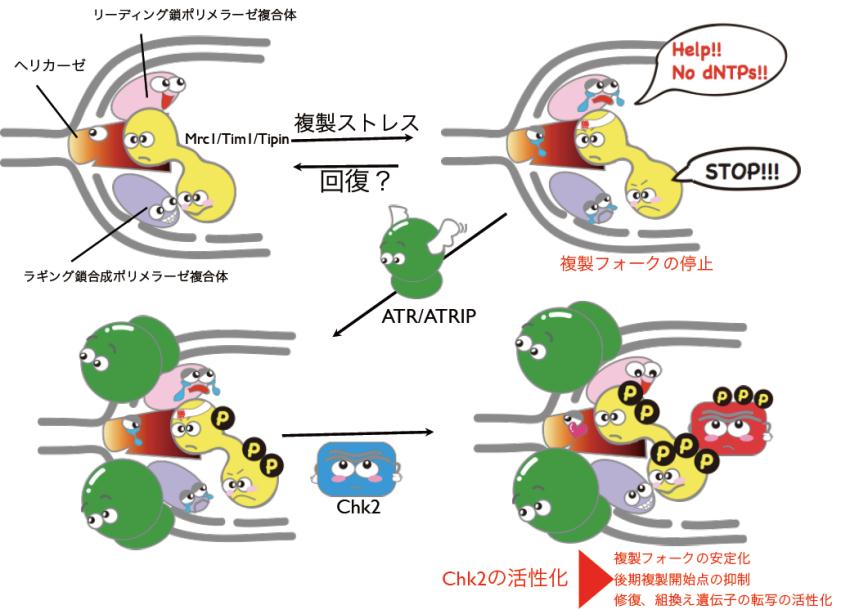
Fig2 ChIP-chip analysis reveals checkpoint protein as a component of DNA replication (Katou et al. nature, 2003)

Fig3 Cohesin (ring molecule) and CTCF (orange molecule) co-operates to function as a transcriptional insulator (nature 2008).
This clay model was introduced in News & Views column of nature.
Next generation sequener (NGS) technology has changed our way of performing genomic analyses dramatically. These days ChIP-seq (chromatin immunoprecipitation on DNA sequencer) is getting more popular than ChIP-chip and the technology enables us to explore 90% of human whole genome sequence including majority of repetitive sequences. This is a great advance in our field because by DNA micro array, we could only explore unique sequence on human genome. By utilizing the advantage of this technology at maximum degree, we are now analyzing localization of human specific chromosome associated factors that are previously difficult to analyze by ChIP-chip. Furthermore by applying the emerging technology, Hi-C or ChIA-PET, that allows us to study the high order chromatin structure, now time comes to answer a long standing question “How is all of that DNA packaged so tightly into chromosomes and squeezed into a tiny nucleus?”
The second project is to analyze the function of SMC (Structural Maintenance of Chromosomes) proteins in vertebrate. SMC proteins are classified in 6 sub-families(SMC1 to SMC6). SMC1 and SMC3 are subunits of cohesin complex and play crucial roles for chromosome partition and transcription. SMC2 and SMC4 are subunits of condensing and are essential for chromosome condensation. SMC5 and SMC6 are thought to regulate chromosome topology but detail is not clear at the moment. We have tried to get clues for function of these proteins in vivo through protein binding profile analyses. Our main focus now is to solve the relationships between SMC proteins and transcription.
The third project is chromosome informatics. We are extending our ChIP-seq analyses, revealing localization profiles of more than 300 DNA-binding proteins on S. cerevisiae, S. pombe, and human chromosomes. These profile maps will tell us not only about molecular basis of functional elements on eukaryotic chromosomes(i.e. replication origins, cohesin sites, centromeres and so on), but also on how these functional elements are organized to construct the dynamic and integrated chromosome architecture. Moreover, comparison of chromosome structures of two yeast species, which have very different genome structures despite their similarity in overall genome sizes, would lead us to realizing what becomes more crucial to maintain longer and more complex genomes like human chromosomes. We are also developing software to handle, analyze, and visualize huge ChIP-seq data sets easily and intuitively.


