Research
The Chromatin
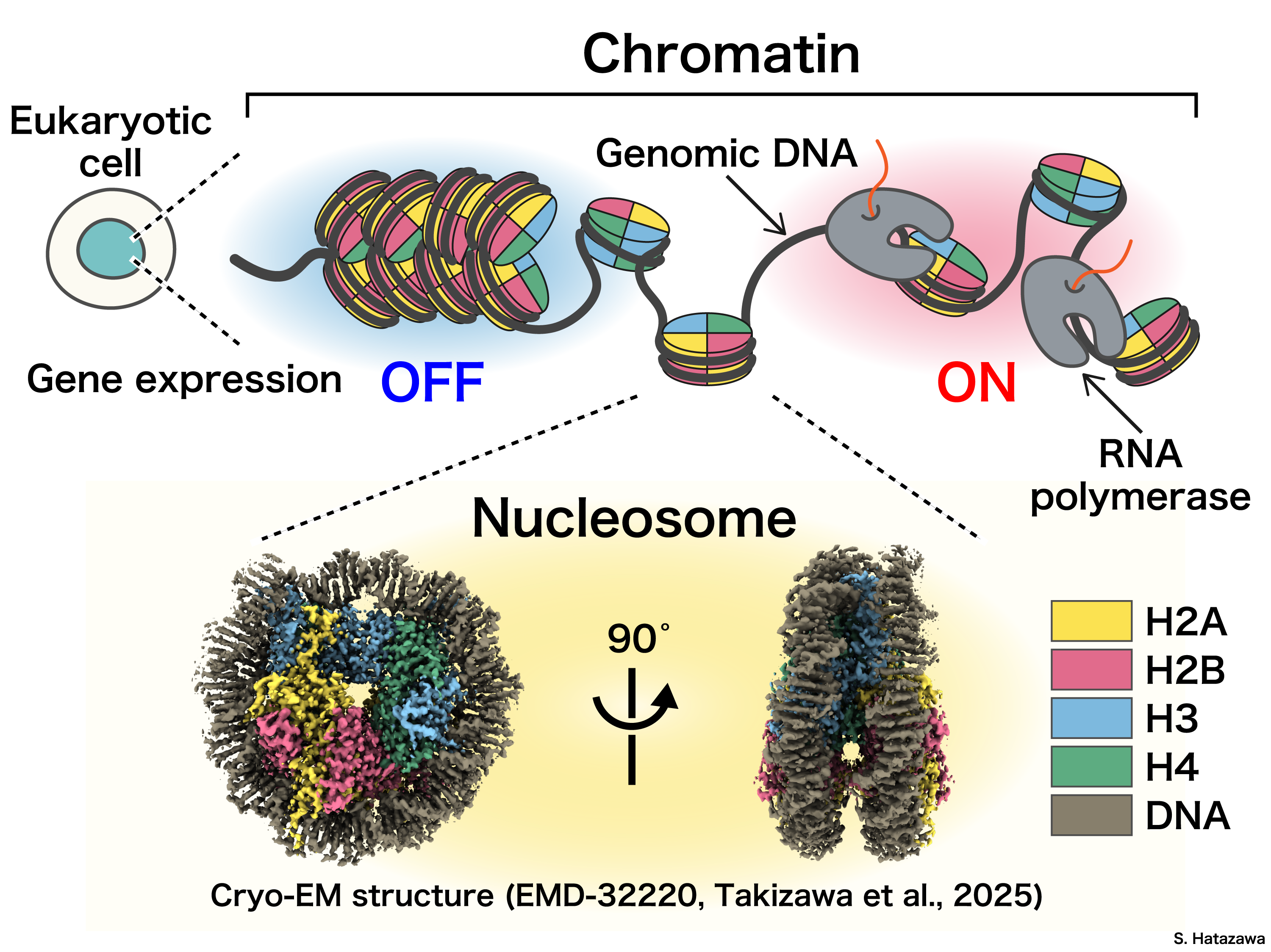
As widely known, our genetic information is encoded in DNA sequences and stored in the chromatin. However, DNA sequences alone seem not the explain to all traits we have – if you are familiar with the fact that all cells in our body originate from a single fertilized egg, which means they have nearly identical DNA sequences. How do cells’ identities get defined in early developmental and differentiation processes? How can cells function differently with same set of genetic information? It is clear there must be some set of regulatory mechanisms operating gene expression without altering the DNA sequence. To investigate such mechanism, in our lab we focus on this field: epigenetics.
Chromatin structure is proven playing a pivotal role in understanding the mechanisms of epigenetic regulation. For instance, structural changes in chromatin, such as by histone variants/modifications or chromatin remodeling, are known to influence gene expression. The molecular mechanisms by which these structural changes regulate gene expression have earned significant attention in recent years.
Our research aims to uncover the molecular mechanisms underlying epigenetic regulation. As structural biologists, we are interested in how structural changes in chromatin drive gene expression by directly visualizing chromatin and analyzing its architecture and function. To achieve so, we strived for years and have established highly realizable core techniques in our lab, which are introduced in detail in following sections.
Reconstitution of Chromatin in vitro
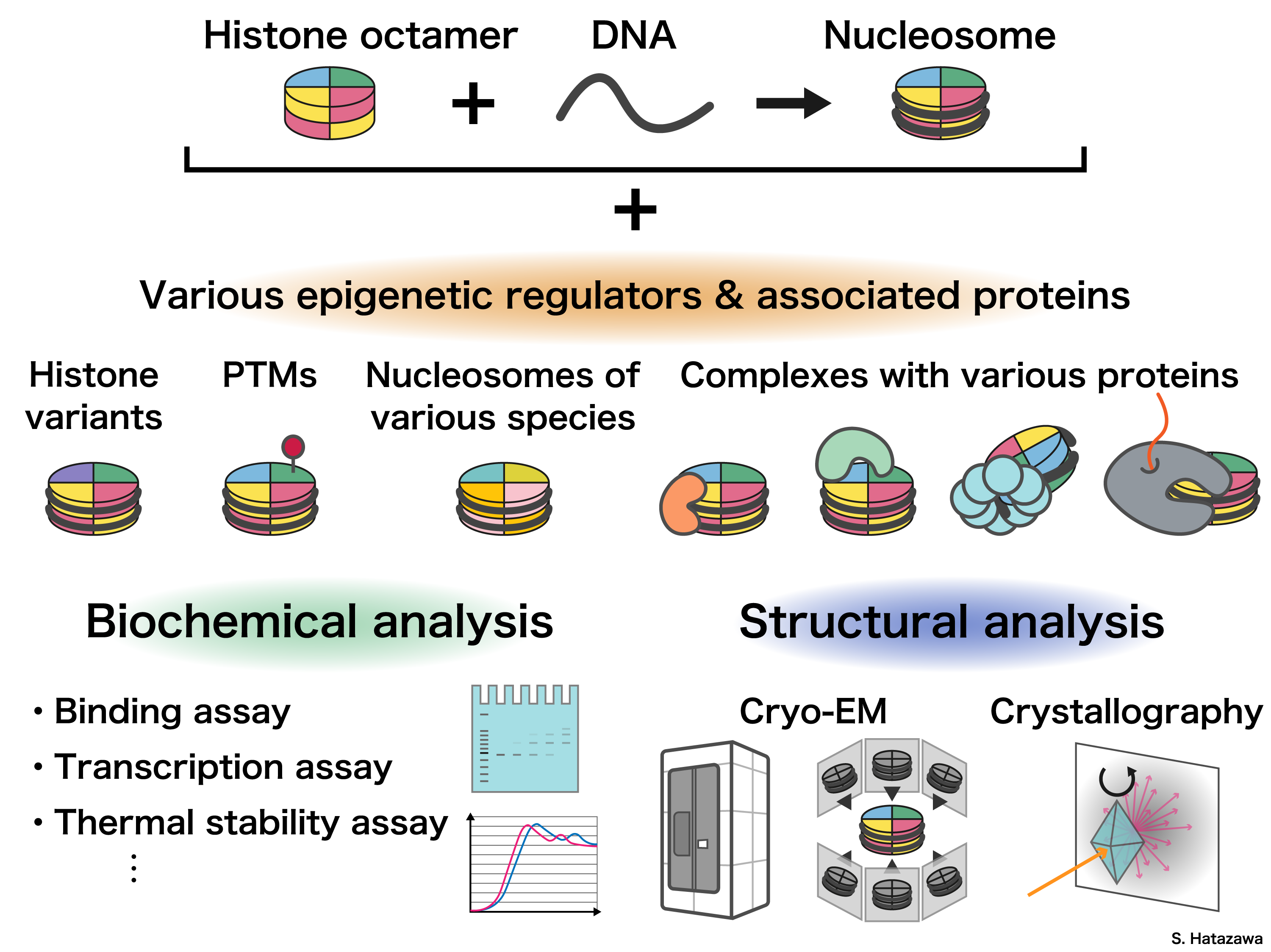
To study chromatin structure, we need first understand better on its fundamental unit: nucleosome. It consists of DNA wrapped around a core of four histone proteins: H2A, H2B, H3, and H4, forming a compact structure. Exposed in the nuclear environment, chromatin interacts with a wide variety of proteins and RNAs, which play crucial roles in regulating its structure and function.
We purify DNA and proteins to reconstitute chromatin in vitro. This technique allows us to reconstitute specific chromatin structures that are challenging to isolate directly from cells. By analyzing the structure and characteristics of these reconstituted chromatin, we aim to gain insights into their functions.
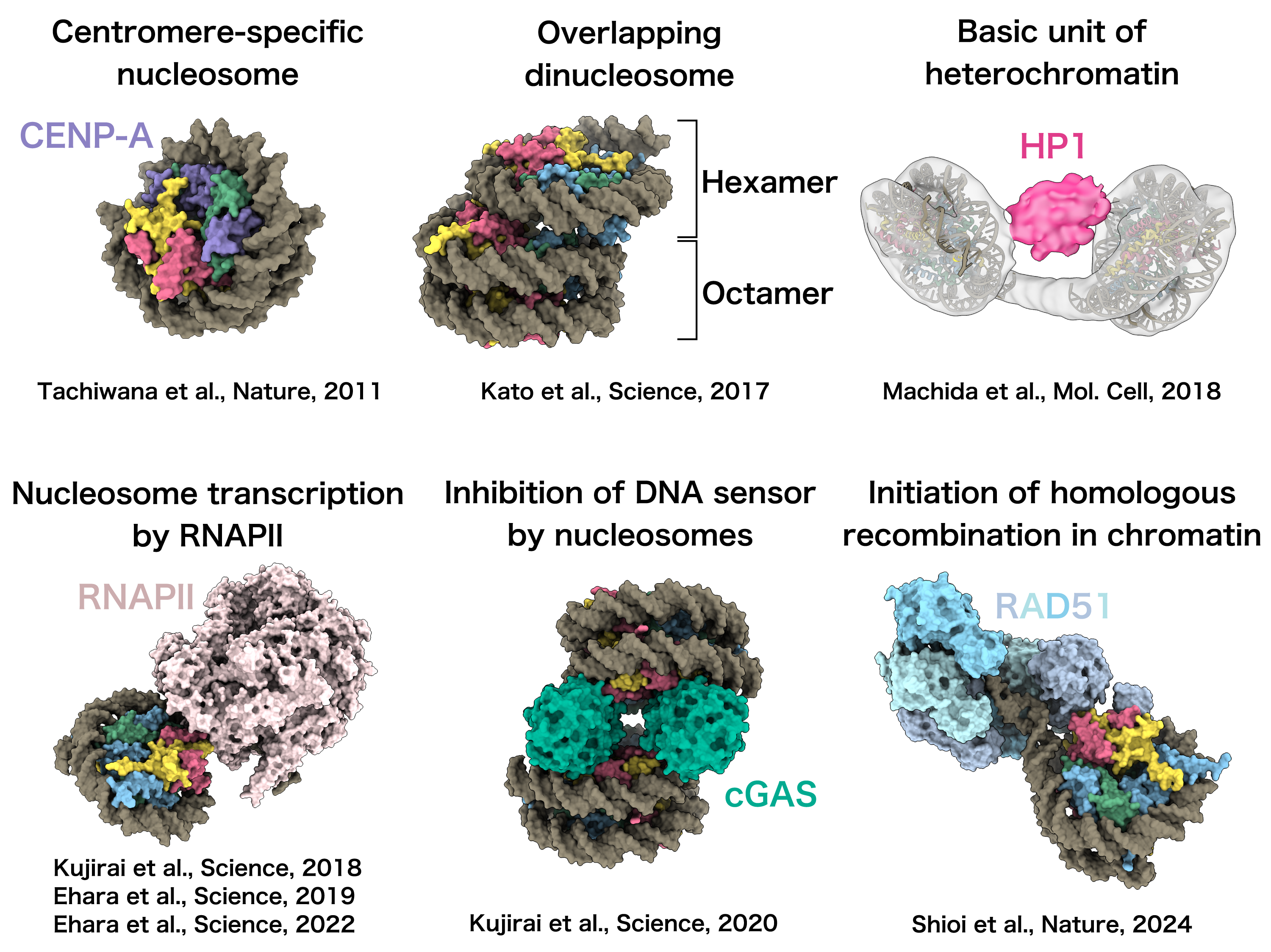
Using cryo-electron microscopy (cryo-EM) to analyze reconstituted chromatin, we have elucidated the molecular mechanisms underlying various biological processes occurring on chromatin.
For example, our studies have revealed:
• The structure of the centromere-specific non-canonical nucleosome, a unique unit of chromatin;
• The architecture of nucleosomes transcribed by RNA polymerase II;
• The mechanism of DNA sensing inhibited by nucleosomes;
• The initiation steps of homologous recombination on chromatin.
These findings provide critical insights into the structural and functional dynamics of chromatin in key biological processes.
Extracting Chromatin from Cells
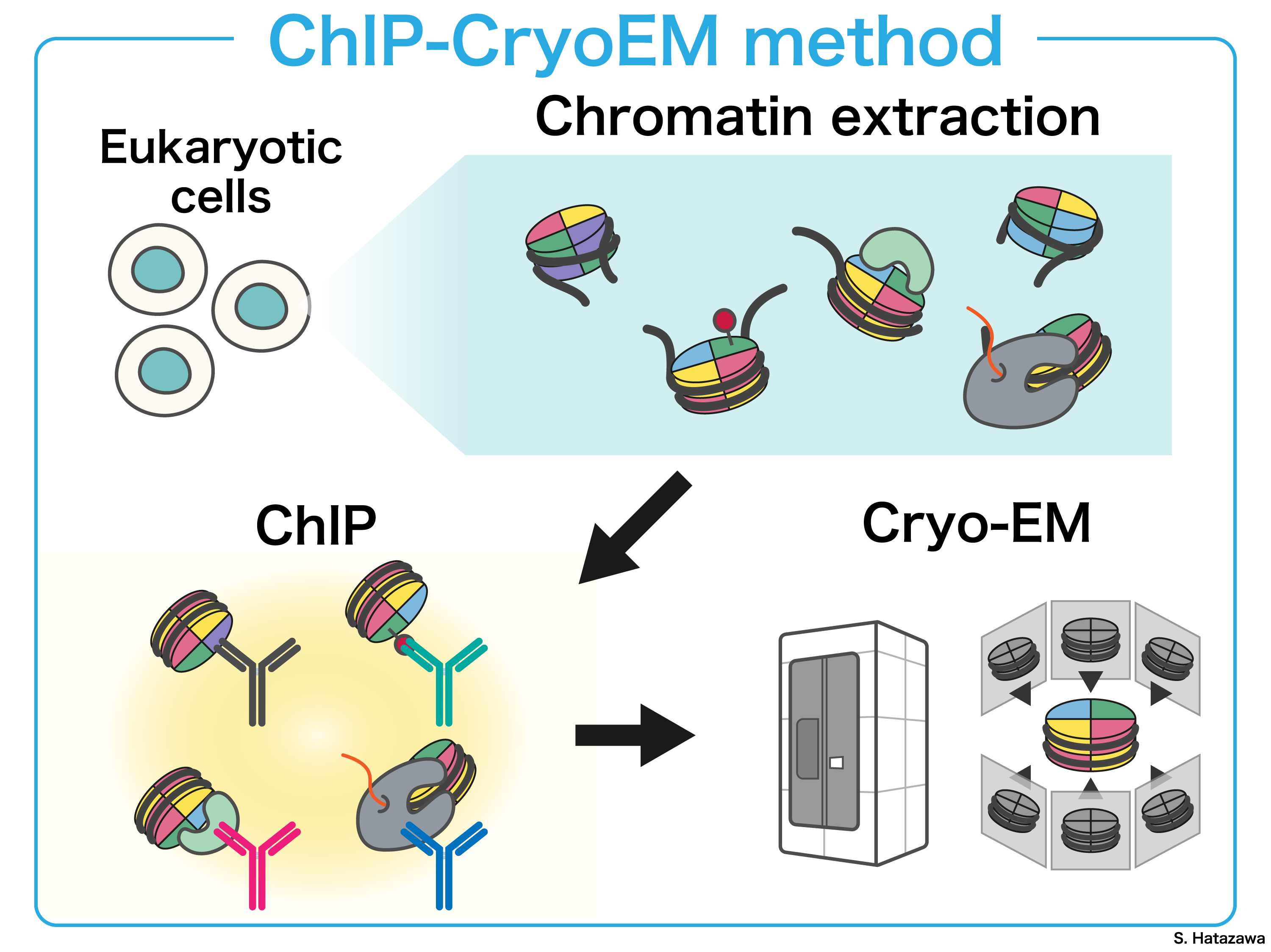
In the cell nucleus, chromatin structure dynamics is precisely orchestrated by the interplay of various epigenomic factors, including histone variants, post-translational modifications (PTMs) of histones, and chromatin-associated proteins. By selectively extracting chromatin containing epigenomic factors of interest from cells, we can further investigate its structural features in detail. This research direction enables us gaining deeper insights into the molecular mechanisms underlying various nuclear functions occurring in cell.
In short, our goal is to establish a comprehensive set of structural analysis methodologies for chromatin isolated through immunoprecipitation containing diverse epigenomic factors. One such technology we have developed is ChIP-CryoEM. Ultimately, we aim to deepen our understanding of nuclear phenomena, through structural insights gained from extracted chromatin.
Analyzing Chromatin in situ
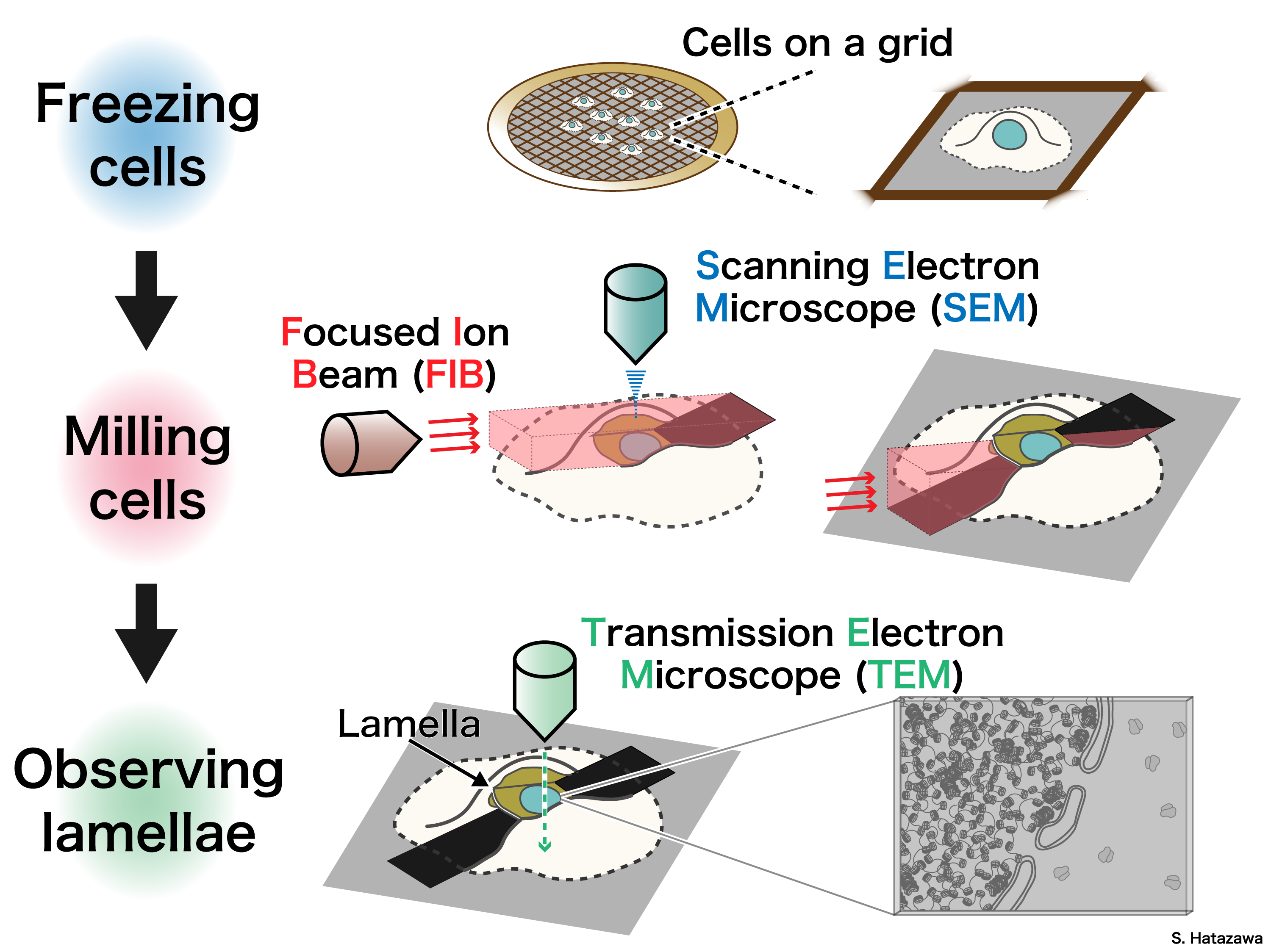
In addition to in vitro experiments, we aim to further investigate how chromatin regulates the functions of genomic DNA within the cellular environment to capture chromatin dynamics in a more physiologically relevant context. We employ cryo-electron tomography to directly analyze chromatin in the nucleus.
Using focused ion beam milling, we prepare ultrathin sections of vitrified cells, making them suitable for analysis by transmission electron microscopy. By capturing tilted images of these cell sections, we reconstruct the three-dimensional structures of chromatin in situ, providing valuable insights into its native organization and dynamics.
